Performance of Subgenomic RT-PCR for Predicting SARS-CoV-2 Infectivity Compared to Genomic RT-PCR and Culture Isolation
- PMID: 40272020
- PMCID: PMC12020333
- DOI: 10.1002/jmv.70363
Performance of Subgenomic RT-PCR for Predicting SARS-CoV-2 Infectivity Compared to Genomic RT-PCR and Culture Isolation
Abstract
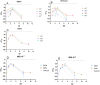
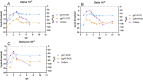
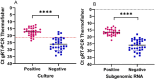
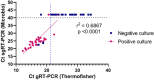
SARS-CoV-2 clinical samples can be detected as positive for a long period of time using real-time RT-PCR, even when patients are no longer infectious. Viral culture is the gold standard for assessing a patient's infectivity, but it is a time-consuming technique and lacks sensitivity. SARS-CoV-2 subgenomic RNA (sgRNA) detection has been used as a proxy for assessing the infectivity but only a limited number of studies have described its use in vitro and in clinical samples. This study aimed to evaluate the correlation between results from viral culture, genomic RT-PCR (gRT-PCR), and subgenomic RT-PCR (sgRT-PCR) during in vitro infection and in clinical samples. In vitro viral replication kinetics showed that both genomic RNA (gRNA) and subgenomic RNA (sgRNA) levels remained stable up to 21 days in the absence of replication-competent virus. Using clinical samples, sgRNA was detected in 87.5% of culture-positive samples, demonstrating better performances than gRT-PCR (Positive predictive value (PPV) 93.3% and Negative predictive value (NPV) of 87.5%) and an almost perfect agreement with culture results (Cohen κ = 0.81 [95% CI: 0.66-0.95]). These findings suggest that testing for sgRNA and/or using a gRNA Ct cut-off of 21.2 could be used as a proxy to determine the presence of SARS-CoV-2 replication-competent virus.
Keywords: SARS‐CoV‐2; culture; infectivity evaluation; subgenomic RNA; viral isolation.
© 2025 The Author(s). Journal of Medical Virology published by Wiley Periodicals LLC.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
[Evaluation of Viral Subgenomic RNAs and Antigen Presence in SARS-CoV-2 PCR Positive Cases].Mikrobiyol Bul. 2024 Jul;58(3):309-320. doi: 10.5578/mb.20240037. Mikrobiyol Bul. 2024. PMID: 39046212 Turkish.
-
Comparison of the Ct-values for genomic and subgenomic SARS-CoV-2 RNA reveals limited predictive value for the presence of replication competent virus.J Clin Virol. 2023 Aug;165:105499. doi: 10.1016/j.jcv.2023.105499. Epub 2023 May 29. J Clin Virol. 2023. PMID: 37327554 Free PMC article.
-
Severe Acute Respiratory Syndrome Coronavirus 2 Normalized Viral Loads and Subgenomic RNA Detection as Tools for Improving Clinical Decision Making and Work Reincorporation.J Infect Dis. 2021 Oct 28;224(8):1325-1332. doi: 10.1093/infdis/jiab394. J Infect Dis. 2021. PMID: 34329473 Free PMC article.
-
Viral Culture Confirmed SARS-CoV-2 Subgenomic RNA Value as a Good Surrogate Marker of Infectivity.J Clin Microbiol. 2022 Jan 19;60(1):e0160921. doi: 10.1128/JCM.01609-21. Epub 2021 Oct 20. J Clin Microbiol. 2022. PMID: 34669457 Free PMC article.
-
Sensitivity of ID NOW and RT-PCR for detection of SARS-CoV-2 in an ambulatory population.Elife. 2021 Apr 20;10:e65726. doi: 10.7554/eLife.65726. Elife. 2021. PMID: 33876726 Free PMC article.
References
-
- Qutub M., Aldabbagh Y., Mehdawi F., et al., “Duration of Viable SARS‐CoV‐2 Shedding From Respiratory Tract in Different Human Hosts and Its Impact on Isolation Discontinuation Polices Revision; a Narrative Review,” Clinical Infection in Practice 13 (2022): 100140, 10.1016/j.clinpr.2022.100140. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

