Population and pan-genomic analyses of Staphylococcus pseudintermedius identify geographic distinctions in accessory gene content and novel loci associated with AMR
- PMID: 40272117
- PMCID: PMC12094015
- DOI: 10.1128/aem.00010-25
Population and pan-genomic analyses of Staphylococcus pseudintermedius identify geographic distinctions in accessory gene content and novel loci associated with AMR
Abstract
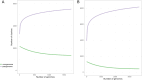
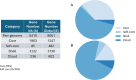
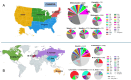
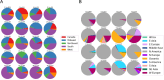
Staphylococcus pseudintermedius is a common representative of the normal skin microbiota of dogs and cats but is also a causative agent of a variety of infections. Although primarily a canine/feline bacterium, recent studies suggest an expanded host range including humans. This paper details population genomic analyses of the largest yet assembled and sequenced collection of S. pseudintermedius isolates from across the USA and Canada and assesses these isolates within a larger global population genetic context. We then employ a pan-genome-wide association study analysis of over 1,700 S. pseudintermedius isolates from sick dogs and cats, covering the period 2017-2020, correlating loci at a genome-wide level, with in vitro susceptibility data for 23 different antibiotics. We find no evidence from either core genome phylogenies or accessory genome content for separate lineages colonizing cats or dogs. Some core genome geographic clustering was evident on a global scale, and accessory gene content was noticeably different between various regions, some of which could be linked to known antimicrobial resistance (AMR) loci for certain classes of antibiotics (e.g., aminoglycosides). Analysis of genes correlated with AMR was divided into different categories, depending on whether they were known resistance mechanisms, on a plasmid, or a putatively novel resistance mechanism on the chromosome. We discuss several novel chromosomal candidates for follow-up laboratory experimentation, including, for example, a bacteriocin (subtilosin), for which the same protein from Bacillus subtilis has been shown to be active against Staphylococcus aureus infections, and for which the operon, present in closely related Staphylococcus species, is absent in S. aureus.IMPORTANCEStaphylococcus pseudintermedius is an important causative agent of a variety of canine and feline infections, with recent studies suggesting an expanded host range, including humans. This paper presents global population genomic data and analysis of the largest set yet sequenced for this organism, covering the USA and Canada as well as more globally. It also presents analysis of in vitro antibiotic susceptibility testing results for the North American (NA) isolates, as well as genetic analysis for the global set. We conduct a pan-genome-wide association study analysis of over 1,700 S. pseudintermedius isolates from sick dogs and cats from NA to correlate loci at a genome-wide level with the in vitro susceptibility data for 23 different antibiotics. We discuss several chromosomal loci arising from this analysis for follow-up laboratory experimentation. This study should provide insight regarding the development of novel molecular treatments for an organism of both veterinary and, increasingly, human medical concern.
Keywords: Staphylococcus pseudintermedius; antibiotic resistance.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






References
-
- Sawhney SS, Vargas RC, Wallace MA, Muenks CE, Lubbers BV, Fritz SA, Burnham C-A, Dantas G. 2023. Diagnostic and commensal Staphylococcus pseudintermedius genomes reveal niche adaptation through parallel selection of defense mechanisms. Nat Commun 14:7065. doi:10.1038/s41467-023-42694-5 - DOI - PMC - PubMed
MeSH terms
Substances
Supplementary concepts
Grants and funding
- U18 FD006862/FD/FDA HHS/United States
- U18 FD006453/FD/FDA HHS/United States
- 1U18FD006558/U.S. Food and Drug Administration
- 1U18FD006862, 1U18FD007244/U.S. Food and Drug Administration
- 1U18FD006442/U.S. Food and Drug Administration
- 1U18FD006453/U.S. Food and Drug Administration
- 1U18FD006993/U.S. Food and Drug Administration
- U18 FD006712/FD/FDA HHS/United States
- 5U18FD006716/U.S. Food and Drug Administration
- 1U18FD006712/U.S. Food and Drug Administration
- U18 FD007244/FD/FDA HHS/United States
- U18 FD006993/FD/FDA HHS/United States
- U18 FD006442/FD/FDA HHS/United States
- U18 FD006558/FD/FDA HHS/United States
- U18 FD006716/FD/FDA HHS/United States
- AP20VSD&B000C018/U.S. Department of Agriculture
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

