Structural and functional significance of Aedes aegypti AgBR1 flavivirus immunomodulator
- PMID: 40272158
- PMCID: PMC12090808
- DOI: 10.1128/jvi.01878-24
Structural and functional significance of Aedes aegypti AgBR1 flavivirus immunomodulator
Abstract
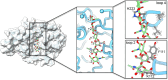
Zika virus (ZIKV), an arbovirus, relies on mosquitoes as vectors for its transmission. During blood feeding, mosquitoes inoculate saliva containing various proteins. Recently, AgBR1, an Aedes aegypti salivary gland protein, has gained attention for its immunomodulatory potential, along with another protein, called NeSt1. We have determined the crystal structure of AgBR1 at 1.2 Å resolution. Despite its chitinase-like fold, we demonstrated that AgBR1 does not bind to chitobiose or chitinhexaose, while a key mutation in the catalytic site abrogates enzymatic activity, suggesting that the protein's function has been repurposed. Our study also shows that AgBR1 and NeSt1, when presented to murine primary macrophages, alter cellular pathways related to virus entry by endocytosis, immune response, and cell proliferation. AgBR1 (and NeSt1) do not directly bind to the Zika virus or modulate its replication. We propose that their immunomodulatory effects on Zika virus transmission are through regulation of host-cell response, a consequence of evolutionary cross talk and virus opportunism. These structural and functional insights are prerequisites for developing strategies to halt the spread of mosquito-borne disease.IMPORTANCEOur study informs on the structural and functional significance of a mosquito salivary gland protein, AgBR1 (along with another protein called NeSt1), in the transmission of the Zika virus (ZIKV), a mosquito-borne virus that has caused global health concerns. By analyzing AgBR1's three-dimensional structure in combination with cellular and interaction studies, we discovered that AgBR1 does not function like typical proteins in its family-it does not degrade sugars. However, we show that it primes immune cells in a way that could help the virus enter cells more easily but not by interacting with the virus or altering viral replication. This finding is significant because it reveals how mosquito proteins, repurposed by evolution, can influence virus transmission without the virus's direct presence. Understanding how proteins like AgBR1 work could guide the development of new strategies to prevent Zika virus spread, with potential relevance for other mosquito-borne viruses.
Keywords: Zika virus; cellular pathways; crystal structure; immunomodulators; mosquito salivary proteins; mosquito-borne disease.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Aedes aegypti NeSt1 Protein Enhances Zika Virus Pathogenesis by Activating Neutrophils.J Virol. 2019 Jun 14;93(13):e00395-19. doi: 10.1128/JVI.00395-19. Print 2019 Jul 1. J Virol. 2019. PMID: 30971475 Free PMC article.
-
Aedes aegypti AgBR1 antibodies modulate early Zika virus infection of mice.Nat Microbiol. 2019 Jun;4(6):948-955. doi: 10.1038/s41564-019-0385-x. Epub 2019 Mar 11. Nat Microbiol. 2019. PMID: 30858571 Free PMC article.
-
AgBR1 and NeSt1 antisera protect mice from Aedes aegypti-borne Zika infection.Vaccine. 2021 Mar 19;39(12):1675-1679. doi: 10.1016/j.vaccine.2021.01.072. Epub 2021 Feb 20. Vaccine. 2021. PMID: 33622591 Free PMC article.
-
Zika Virus Mosquito Vectors: Competence, Biology, and Vector Control.J Infect Dis. 2017 Dec 16;216(suppl_10):S976-S990. doi: 10.1093/infdis/jix405. J Infect Dis. 2017. PMID: 29267910 Free PMC article. Review.
-
Mosquito-borne and sexual transmission of Zika virus: Recent developments and future directions.Virus Res. 2018 Aug 2;254:1-9. doi: 10.1016/j.virusres.2017.07.011. Epub 2017 Jul 11. Virus Res. 2018. PMID: 28705681 Free PMC article. Review.
References
-
- Jin L, Guo X, Shen C, Hao X, Sun P, Li P, Xu T, Hu C, Rose O, Zhou H, Yang M, Qin CF, Guo J, Peng H, Zhu M, Cheng G, Qi X, Lai R. 2018. Salivary factor LTRIN from Aedes aegypti facilitates the transmission of Zika virus by interfering with the lymphotoxin-β receptor. Nat Immunol 19:342–353. doi:10.1038/s41590-018-0063-9 - DOI - PubMed
MeSH terms
Substances
Grants and funding
- R41 AI152904/AI/NIAID NIH HHS/United States
- AI152904/HHS | NIH | All of Us Research Program (All of Us NIH)
- RTI2018-095700-B-I00, PID2021-126130OB-I00/Agencia Estatal de Investigación
- RTI2018-094751-B-C21, PDI2021-1237810B-C21, PID2021-124328OB-I00/Agencia Estatal de Investigación
- WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Medical

