Comparative genomic analysis of metal-tolerant bacteria reveals significant differences in metal adaptation strategies
- PMID: 40272196
- PMCID: PMC12131726
- DOI: 10.1128/spectrum.01680-24
Comparative genomic analysis of metal-tolerant bacteria reveals significant differences in metal adaptation strategies
Abstract
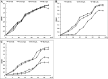
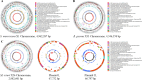
Metal-tolerant bacteria have been commercially used in wastewater treatment, bio-fertilizer, and soil remediation, etc. However, the mechanisms underlying their actions are not yet fully understood. We isolated metal-tolerant bacteria from the rhizosphere soil samples with metal-enriched media containing Cu, Fe, or Mn, sequenced and compared the genomes, and analyzed their metal adaptation strategies at genomic levels to better understand their action mechanisms. Totally, 32 metal-tolerant isolates were identified and classified into 12 genera based on phylogenetic analysis. The determination of maximum tolerance concentration and the effect of metal ions on the isolates indicated that Serratia marcescens X1 (CuSO4: 1,000 mg/L, FeSO4: 1,000 mg/L, and MnSO4.4H2O: 2,000 mg/L), Mammaliicoccus sciuri X26 (FeSO4: 600 mg/L and MnSO4.4H2O: 2,000 mg/L), and Rummeliibacillus pycnus X33 (CuSO4: 400 mg/L, FeSO4: 1,000 mg/L, and MnSO4.4H2O: 800 mg/L) showed significant differences in metal tolerance to Cu, Fe, and Mn with other isolates. They possess quite different genomic features that enable them to adapt to various metal ions. S. marcescens X1 possesses abundant genes required for Cu, Fe, and Mn homeostasis. M. sciuri X26 has a number of genes involved in Mn and Zn homeostasis but with no genes responsible for Cu and Ca transport. R. pycnus X33 is rich in Fe, Zn, and Mg transport systems but poor in Cu and Mn transport systems. It is thus inferred that the combined use of them would compensate for their differences and enhance their ability in accumulating a wider range of heavy metals for promoting their applications in industry, agriculture, and ecology.
Importance: Metal-tolerant bacteria have wide applications in environmental, agricultural, and ecological fields, but their action strategies are not yet fully understood. We isolated 32 metal-tolerant bacteria from the rhizosphere soil samples. Among them, Serratia marcescens X1, Mammaliicoccus sciuri X26, and Rummeliibacillus pycnus X33 showed significant differences in metal tolerance to Cu, Fe, and Mn with other isolates. Comparative genomic analysis revealed that they have abundant and different genomic features to adapt to various metal ions. It is thus inferred that the combined use of them would compensate for their differences and enhance their ability to accumulate heavy metal ions, widening their applications in industry, agriculture, and ecology.
Keywords: application; combined use; genomic analysis; metal-stress-response; metal-tolerant bacteria.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Characterization and bioremediation potential of native heavy-metal tolerant bacteria isolated from rat-hole coal mine environment.Arch Microbiol. 2021 Jul;203(5):2379-2392. doi: 10.1007/s00203-021-02218-5. Epub 2021 Mar 4. Arch Microbiol. 2021. PMID: 33665708
-
Effects of chicken and pig manures application on heavy metal in different soil types and on bacterial in the rhizosphere soil of Chinese cabbage.Antonie Van Leeuwenhoek. 2025 Apr 26;118(6):75. doi: 10.1007/s10482-025-02085-w. Antonie Van Leeuwenhoek. 2025. PMID: 40285902
-
Analysis of copper tolerant rhizobacteria from the industrial belt of Gujarat, western India for plant growth promotion in metal polluted agriculture soils.Ecotoxicol Environ Saf. 2017 Apr;138:113-121. doi: 10.1016/j.ecoenv.2016.12.023. Epub 2016 Dec 28. Ecotoxicol Environ Saf. 2017. PMID: 28038338
-
Characterization of copper-resistant bacteria and bacterial communities from copper-polluted agricultural soils of central Chile.BMC Microbiol. 2012 Sep 5;12:193. doi: 10.1186/1471-2180-12-193. BMC Microbiol. 2012. PMID: 22950448 Free PMC article.
-
Implications of metal accumulation mechanisms to phytoremediation.Environ Sci Pollut Res Int. 2009 Mar;16(2):162-75. doi: 10.1007/s11356-008-0079-z. Epub 2008 Dec 6. Environ Sci Pollut Res Int. 2009. PMID: 19067014 Review.
References
Publication types
MeSH terms
Substances
Grants and funding
- 2020GDASYL-20200102009/Guangdong Academy of Sciences
- 2022GDASZH-2022010110/Guangdong Academy of Sciences
- 2020GDASYL-20200102010/Guangdong Academy of Sciences
- 2021CX020301/Guangdong Provincial Department of Science and Technology (GDSTC)
- 2022A1515110562/Basic and Applied Basic Research Foundation of Guangdong Province
LinkOut - more resources
Full Text Sources

