Accelerating Medicines Partnership in Type 2 Diabetes and Common Metabolic Diseases: Collaborating to Maximize the Value of Genetic and Genomic Data
- PMID: 40272257
- PMCID: PMC12185966
- DOI: 10.2337/db25-0042
Accelerating Medicines Partnership in Type 2 Diabetes and Common Metabolic Diseases: Collaborating to Maximize the Value of Genetic and Genomic Data
Abstract
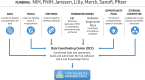
In the last two decades, significant progress has been made toward understanding the genetic basis of type 2 diabetes. An important supporter of this research has been the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK), most recently through the Accelerating Medicines Partnership Program for Type 2 Diabetes (AMP T2D) and Accelerating Medicines Partnership Program for Common Metabolic Diseases (AMP CMD). These public-private partnerships of the National Institutes of Health, multiple biopharmaceutical and life sciences companies, and nonprofit organizations, facilitated and managed by the Foundation for the National Institutes of Health, were designed to improve understanding of therapeutically relevant biological pathways for type 2 diabetes. On the occasion of NIDDK's 75th anniversary, we review the history of NIDDK support for these partnerships, which saw the convergence of research directions prioritized by academic consortia, the pharmaceutical industry, and government funders. Although the NIDDK was not the sole originator or funder of these efforts, its support and leadership have been pivotal to the partnerships' success and have enabled their research to be broadly accessible through the AMP Common Metabolic Diseases Knowledge Portal (CMDKP) and the AMP Common Metabolic Diseases Genome Atlas (CMDGA). Findings from AMP CMD align with NIDDK's mission to conduct research and share results with the goal of improving health and quality of life.
Article highlights: The Accelerating Medicines Partnership Program for Type 2 Diabetes (AMP T2D) and Accelerating Medicines Partnership Program for Common Metabolic Diseases (AMP CMD) were created to accelerate the translation of genetic and genomic data into knowledge about the biology of disease. Their goal was to gain a better understanding of the mechanisms underlying types 1 and 2 diabetes and prediabetes, obesity, cardiovascular disease, kidney disease, and nonalcoholic steatohepatitis. This work identified multiple genes and pathways underlying these diseases. The findings of AMP T2D and AMP CMD have implications for drug development and improved risk prediction, diagnosis, and treatment for common metabolic diseases.
© 2025 by the American Diabetes Association.
Conflict of interest statement
Figures


References
-
- Diabetes Genetics Initiative of Broad Institute of Harvard and MIT, Lund University, and Novartis Institutes of BioMedical Research ; Saxena R, Voight BF, Lyssenko V, et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science 2007;316:1331–1336 - PubMed
Publication types
MeSH terms
Grants and funding
- UM1 DK126194/DK/NIDDK NIH HHS/United States
- R01DK072193/DK/NIDDK NIH HHS/United States
- UM1 DK105554/DK/NIDDK NIH HHS/United States
- U01 DK085545/DK/NIDDK NIH HHS/United States
- MR/L020149/1/MRC_/Medical Research Council/United Kingdom
- R01 DK072193/DK/NIDDK NIH HHS/United States
- R01 DK125497/DK/NIDDK NIH HHS/United States
- R01DK093757/DK/NIDDK NIH HHS/United States
- UM1DK126194/DK/NIDDK NIH HHS/United States
- 095101/WT_/Wellcome Trust/United Kingdom
- R01HD056465/National Institute of Child Health and Human Development
- 098381/WT_/Wellcome Trust/United Kingdom
- U01 DK123594/DK/NIDDK NIH HHS/United States
- NNF21SA0072102/Novo Nordisk Fonden
- U01DK105535/DK/NIDDK NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- UM1 DK126185/DK/NIDDK NIH HHS/United States
- 106130/WT_/Wellcome Trust/United Kingdom
- R01 DK093757/DK/NIDDK NIH HHS/United States
- U01DK123594/DK/NIDDK NIH HHS/United States
- U01DK134995/DK/NIDDK NIH HHS/United States
- 200837/WT_/Wellcome Trust/United Kingdom
- R01 DK062370/DK/NIDDK NIH HHS/United States
- 212259/WT_/Wellcome Trust/United Kingdom
- U01 DK105535/DK/NIDDK NIH HHS/United States
- UM1DK105554/DK/NIDDK NIH HHS/United States
- R01 HD056465/HD/NICHD NIH HHS/United States
- U01DK085545/DK/NIDDK NIH HHS/United States
- 090532/WT_/Wellcome Trust/United Kingdom
- UM1DK126185/DK/NIDDK NIH HHS/United States
- R01DK125497/DK/NIDDK NIH HHS/United States
- 203141/WT_/Wellcome Trust/United Kingdom
- R01DK062370/DK/NIDDK NIH HHS/United States
- U01 DK134995/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical

