Identifying Four Obesity Axes Through Integrative Multiomics and Imaging Analysis
- PMID: 40272846
- PMCID: PMC12185968
- DOI: 10.2337/db24-1103
Identifying Four Obesity Axes Through Integrative Multiomics and Imaging Analysis
Abstract
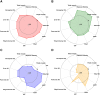
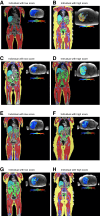
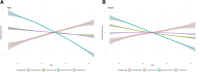
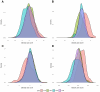
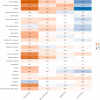
We aimed to identify distinct axes of obesity using advanced magnetic resonance imaging (MRI)-derived phenotypes. We used 24 MRI-derived fat distribution and muscle volume measures (UK Biobank; N = 33,122) to construct obesity axes through principal component analysis. Genome-wide association studies were performed for each axis to uncover genetic factors, followed by pathway enrichment, genetic correlation, and Mendelian randomization analyses to investigate disease associations. Four primary obesity axes were identified: 1) general obesity, reflecting higher fat accumulation in all regions (visceral, subcutaneous, and ectopic fat); 2) muscle dominant, indicating greater muscle volume; 3) peripheral fat, associated with higher subcutaneous fat in abdominal and thigh regions; and 4) lower-body fat, characterized by increased lower-body subcutaneous fat and reduced ectopic fat. Each axis was associated with distinct genetic loci and pathways. For instance, the lower-body fat axis was associated with RSPO3 and COBLL1, which are emerging as promising candidates for therapeutic targeting. Disease risks varied across axes; the general obesity axis was correlated with higher risks of metabolic and cardiovascular diseases, whereas the lower-body fat axis seemed to protect against type 2 diabetes and cardiovascular disease. This study highlights the heterogeneity of obesity through the identification of obesity axes and emphasizes the potential to extend beyond BMI in defining and treating obesity for obesity-related disease management.
Article highlights: This study aimed to address potential limitations of BMI by exploring the heterogeneity of obesity using magnetic resonance imaging-derived fat distribution and muscle volume measures. We sought to identify distinct obesity axes and investigate their genetic, metabolic, and disease associations. Four obesity axes were identified (general obesity, muscle dominant, peripheral fat, and lower-body fat), each linked to unique genetic loci, metabolic traits, and disease risks. These findings emphasize the potential to extend beyond BMI in defining and managing obesity, offering a more nuanced framework for understanding and treating obesity-related diseases.
© 2025 by the American Diabetes Association.
Conflict of interest statement
Figures


References
-
- Abraham A, Yaghootkar H. Identifying obesity subtypes: a review of studies utilising clinical biomarkers and genetic data. Diabet Med 2023;40:e15226. - PubMed
-
- Bray GA. Medical consequences of obesity. J Clin Endocrinol Metab 2004;89:2583–2589 - PubMed
-
- Perdomo CM, Cohen RV, Sumithran P, Clément K, Frühbeck G. Contemporary medical, device, and surgical therapies for obesity in adults. Lancet 2023;401:1116–1130 - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

