Label-Free Quantification of Protein Density in Living Cells
- PMID: 40272941
- PMCID: PMC12021013
- DOI: 10.1002/cpz1.70130
Label-Free Quantification of Protein Density in Living Cells
Abstract
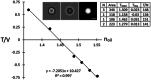
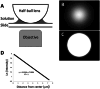
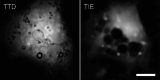
Intracellular water content, W, and protein concentration, P, are essential characteristics of living cells. Healthy cells maintain them within a narrow range, but often become dehydrated under severe stress; moreover, persistent loss of water (an increase in P) can lead to apoptotic death. It is very likely that protein concentration affects cellular metabolism and signaling through macromolecular crowding (MC) effects, to which P is directly related, but much remains unknown in this area. Obviously, in order to study the biological roles and regulation of MC in living cells, one needs a method to measure it. Simple and accurate measurements of P in adherent cells can be based on its relationship to refractive index. The latter can be derived from two or more (depending on the algorithm) mutually defocused brightfield images processed by the transport-of-intensity equation (TIE) that must be complemented by a determination of volume. Here, we describe the experimental considerations for both TIE imaging and for a particular method of cell volume measurement, transmission-through-dye (TTD). We also introduce an ImageJ plugin for solving TIE. TIE and TTD are fully compatible with each other as well as with fluorescence. A similar approach can be applied to subcellular organelles; however, in this case, the volume must be determined differently.© 2025 The Author(s). Current Protocols published by Wiley Periodicals LLC. Basic Protocol 1: Sample preparation for TIE with or without TTD Basic Protocol 2: Acquisition of TIE and TTD images Basic Protocol 3: Calibration of TIE Basic Protocol 4: Measurement of the absorption coefficient of the medium used for TTD Basic Protocol 5: Image processing using Fiji Support Protocol 1: Installation and use of TIE plugin Support Protocol 2: Automation of the double TTD/TIE processing using a Fiji macro.
Keywords: cell volume; macromolecular crowding; protein concentration; refractive index; transmission‐through‐dye microscopy; transport‐of‐intensity equation.
© 2025 The Author(s). Current Protocols published by Wiley Periodicals LLC.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
Literature Cited
-
- Andreas, B. (2025). Simple Transport of Intensity Equation Phase Unwrapper (stiepu.m) (https://www.mathworks.com/matlabcentral/fileexchange/67327‐simple‐transp...) MATLAB Central File Exchange.
-
- Cadart, C. , Zlotek‐Zlotkiewicz, E. , Venkova, L. , Thouvenin, O. , Racine, V. , le Berre, M. , Monnier, S. , & Piel, M. (2017). Fluorescence eXclusion Measurement of volume in live cells. In Methods in Cell Biology (Vol. 139, pp. 103–120). Academic Press. - PubMed
-
- Cayley, S. , Lewis, B. A. , Guttman, H. J. , & Record Jr, M. T. (1991). Characterization of the cytoplasm of Escherichia coli K‐12 as a function of external osmolarity: Implications for protein‐DNA interactions in vivo. Journal of Molecular Biology, 222(2), 281–300. 10.1016/0022-2836(91)90212-O - DOI - PubMed
Internet Resources
-
- An alternative TIE code.
-
- An alternative TIE code.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

