Genomic insights into the taxonomic status and bioactive gene cluster profiling of Bacillus velezensis RVMD2 isolated from desert rock varnish in Ma'an, Jordan
- PMID: 40273114
- PMCID: PMC12021177
- DOI: 10.1371/journal.pone.0319345
Genomic insights into the taxonomic status and bioactive gene cluster profiling of Bacillus velezensis RVMD2 isolated from desert rock varnish in Ma'an, Jordan
Abstract
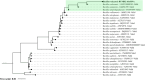
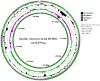
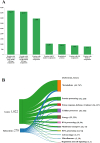
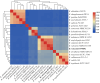
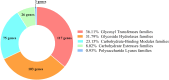
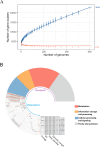
Extreme environments like arid and semi-arid deserts harbor unique microbial diversity, offering rich sources of specialized microbial metabolites. This study explores Bacillus velezensis RVMD2, a strain isolated from rock varnish in the Ma'an Desert, Jordan. The genome was sequenced using the Illumina NextSeq 2000 platform, resulting in a 4,212,579 bp assembly with a GC content of 45.94%. The assembled genome comprises 112 contigs and encodes 4,250 proteins, 77 tRNA genes, and 4 rRNA genes. Phylogenetic analysis of the 16S rRNA gene indicated a 99.84% similarity to previously identified B. velezensis strains. Whole-genome phylogeny using EzBiome, MiGA, and TYGS confirmed its classification as B. velezensis. Functional annotation identified genes involved in carbohydrate metabolism, including 324 carbohydrate-active enzyme (CAZyme) genes, stress response, and secondary metabolite biosynthesis. The genome also contains 50 genes associated with heavy metal resistance and plant growth promotion. Analysis using AntiSMASH identified 12 biosynthetic gene clusters involved in the production of secondary metabolites, including fengycin, surfactin, polyketides, terpenes, and bacteriocins. Notably, several clusters did not match any known sequences, suggesting the presence of potentially novel antimicrobial compounds. The genomic features of RVMD2 highlight its adaptability to extreme environments and its potential for biotechnological applications, including bioremediation and the discovery of novel bioactive metabolites.
Copyright: © 2025 Alnaimat et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




Similar articles
-
Comparative Genome Analysis Reveals Phylogenetic Identity of Bacillus velezensis HNA3 and Genomic Insights into Its Plant Growth Promotion and Biocontrol Effects.Microbiol Spectr. 2022 Feb 23;10(1):e0216921. doi: 10.1128/spectrum.02169-21. Epub 2022 Feb 2. Microbiol Spectr. 2022. PMID: 35107331 Free PMC article.
-
Complete genome analysis and antimicrobial mechanism of Bacillus velezensis GX0002980 reveals its biocontrol potential against mango anthracnose disease.Microbiol Spectr. 2025 Jun 3;13(6):e0268524. doi: 10.1128/spectrum.02685-24. Epub 2025 Apr 16. Microbiol Spectr. 2025. PMID: 40237490 Free PMC article.
-
Bacillus haimaensis sp. nov.: a novel cold seep-adapted bacterium with unique biosynthetic potential.Appl Environ Microbiol. 2025 May 21;91(5):e0245624. doi: 10.1128/aem.02456-24. Epub 2025 Apr 25. Appl Environ Microbiol. 2025. PMID: 40277363 Free PMC article.
-
Bacillus velezensis: A Valuable Member of Bioactive Molecules within Plant Microbiomes.Molecules. 2019 Mar 16;24(6):1046. doi: 10.3390/molecules24061046. Molecules. 2019. PMID: 30884857 Free PMC article. Review.
-
Bacillus velezensis: phylogeny, useful applications, and avenues for exploitation.Appl Microbiol Biotechnol. 2019 May;103(9):3669-3682. doi: 10.1007/s00253-019-09710-5. Epub 2019 Mar 25. Appl Microbiol Biotechnol. 2019. PMID: 30911788 Review.
References
-
- Alnaimat S, Shattal SA, Althunibat O, Alsbou E, Amasha R. Iron (II) and other heavy-metal tolerance in bacteria isolated from rock varnish in the arid region of Al-Jafer Basin, Jordan. Biodiversitas. 2017;18(3):1250–7. doi: 10.13057/biodiv/d180350 - DOI
-
- Fatima A, Abbas M, Nawaz S, Rehman Y, Rehman S ur, Sajid I. Whole genome sequencing (WGS) and genome mining of Streptomyces sp. AFD10 for antibiotics and bioactive secondary metabolites biosynthetic gene clusters (BGCs). Gene Reports. 2024;37:102050. doi: 10.1016/j.genrep.2024.102050 - DOI
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Miscellaneous

