The transcriptional regulator VAL1 promotes Arabidopsis flowering by repressing the organ boundary genes BOP1 and BOP2
- PMID: 40273301
- PMCID: PMC12152480
- DOI: 10.1093/plphys/kiaf160
The transcriptional regulator VAL1 promotes Arabidopsis flowering by repressing the organ boundary genes BOP1 and BOP2
Abstract
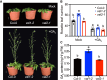
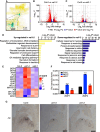
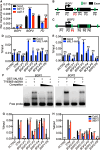
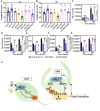
The transition to reproductive development is a critical step in the plant lifecycle and relies on the integration of intrinsic and environmental signals. Several different pathways controlling flowering time function downstream of the perception of environmental cues such as day length (photoperiodic pathway) and seasonal temperature (vernalization and ambient temperature pathways). In addition, the phytohormone gibberellin (GA) induces the floral transition under noninductive photoperiod. In the model plant Arabidopsis (Arabidopsis thaliana), the transcriptional repressor VIVIPAROUS1/ABSCISIC ACID INSENTIVE3 (ABI3)-LIKE1 (VAL1) triggers the stable repression of the floral repressor FLOWERING LOCUS C (FLC) during vernalization. However, the involvement of VAL1 in other flowering pathways remains unclear. In this work, we combined genetic and transcriptomic approaches to investigate the requirement of VAL1 for flowering activation under different day lengths. We found that VAL1, but not its sister protein VAL2, is required to induce the floral transition both under long and short days. The delayed flowering time of val1 mutant plants was fully bypassed by exogenous GA application. We demonstrated that VAL1-mediated induction of flowering occurs partially via the direct epigenetic repression of the organ boundary genes BLADE-ON-PETIOLE1 (BOP1) and BOP2. Our work thus expands the repertoire of VAL target genes and further demonstrates the pleiotropic role of VAL factors in regulating Arabidopsis development.
© The Author(s) 2025. Published by Oxford University Press on behalf of American Society of Plant Biologists.
Conflict of interest statement
Conflict of interest statement. None declared.
Figures


References
-
- Andrés F, Romera-Branchat M, Martínez-Gallegos R, Patel V, Schneeberger K, Jang S, Altmüller J, Nürnberg P, Coupland G. Floral induction in Arabidopsis by FLOWERING LOCUS T requires direct repression of BLADE-ON-PETIOLE genes by the homeodomain protein PENNYWISE. Plant Physiol. 2015:169(3):2187–2199. 10.1104/pp.15.00960 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

