Investigating Plasma Metabolomics and Gut Microbiota Changes Associated With Parkinson Disease: A Focus on Caffeine Metabolism
- PMID: 40273394
- PMCID: PMC12022887
- DOI: 10.1212/WNL.0000000000213592
Investigating Plasma Metabolomics and Gut Microbiota Changes Associated With Parkinson Disease: A Focus on Caffeine Metabolism
Abstract
Background and objectives: Coffee intake is linked to a reduced risk of Parkinson disease (PD), but whether this effect is mediated by gut microbiota and metabolomic changes remains unclear. This study examines PD-associated metabolomic shifts, caffeine metabolism, and their connection to gut microbiome alterations in a multicenter study.
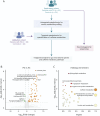
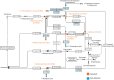
Methods: We conducted an untargeted serum metabolomic assay using liquid chromatography with high-resolution mass spectrometry on an exploratory cohort recruited from National Taiwan University Hospital (NTUH). A targeted metabolomic assay focusing on caffeine and its 12 downstream metabolites was conducted and validated in an independent cohort from University Malaya Medical Centre (UMMC). In the exploratory cohort, the association of each caffeine metabolite with gut microbiota changes was investigated by metagenomic shotgun sequencing. A clustering-based approach was used to correlate microbiome changes with plasma caffeine metabolite level and clinical severity. Body mass index, antiparkinsonism medication use, and dietary habits (including coffee and tea intake) were recorded.
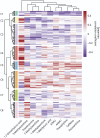
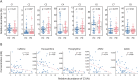
Results: Sixty-three patients with PD and 54 controls from NTUH formed the exploratory cohort while 36 patients with PD and 20 controls from UMMC served as an validation cohort to replicate the plasma caffeine findings. A total of 5,158 metabolites were detected from untargeted metabolomic analysis, with 3,131 having high confidence for analysis. Compared with controls, the abundance of 56 metabolites was significantly higher and that of 7 metabolites was significantly lower (adjusted p < 0.05 and log2 fold change >1) in patients with PD. Caffeine metabolism was significantly lower in patients with PD (p = 0.0013), and serum levels of caffeine and its metabolites negatively correlated with motor severity (p < 0.01). Targeted metabolomic analysis confirmed reduced levels of caffeine and its metabolites, including theophylline, paraxanthine, 1,7-dimethyluric acid, and 5-acetylamino-6-amino-3-methyluracil, in patients with PD; these findings were replicated in the validation cohort (p < 0.05). A clustering approach found that 56 microbiome species enriched in patients with PD negatively correlated with caffeine and its metabolites paraxanthine and theophylline (both p < 0.05), notably Clostridium sp000435655, Acetatifactor sp900066565, Oliverpabstia intestinalis, and Ruminiclostridium siraeum.
Discussion: This study identifies PD-related changes in microbial-caffeine metabolism compared with controls. Our findings offer insights for future functional research on caffeine-microbiome interactions in PD.
Conflict of interest statement
The authors report no relevant disclosures. Go to
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
