Transcriptional regulation by PHGDH drives amyloid pathology in Alzheimer's disease
- PMID: 40273909
- PMCID: PMC12204802
- DOI: 10.1016/j.cell.2025.03.045
Transcriptional regulation by PHGDH drives amyloid pathology in Alzheimer's disease
Abstract
Virtually all individuals aged 65 or older develop at least early pathology of Alzheimer's disease (AD), yet most lack disease-causing mutations in APP, PSEN, or MAPT, and many do not carry the APOE4 risk allele. This raises questions about AD development in the general population. Although transcriptional dysregulation has not traditionally been a hallmark of AD, recent studies reveal significant epigenomic changes in late-onset AD (LOAD) patients. We show that altered expression of the LOAD biomarker phosphoglycerate dehydrogenase (PHGDH) modulates AD pathology in mice and human brain organoids independent of its enzymatic activity. PHGDH has an uncharacterized role in transcriptional regulation, promoting the transcription of inhibitor of nuclear factor kappa-B kinase subunit alpha (IKKa) and high-mobility group box 1 (HMGB1) in astrocytes, which suppress autophagy and accelerate amyloid pathology. A blood-brain-barrier-permeable small-molecule inhibitor targeting PHGDH's transcriptional function reduces amyloid pathology and improves AD-related behavioral deficits. These findings highlight transcriptional regulation in LOAD and suggest therapeutic strategies beyond targeting familial mutations.
Keywords: Alzheimer’s disease; LOAD; PHGDH; amyloid pathology; astrocyte; biomarker; small molecule; therapeutic candidate; transcriptional regulation.
Copyright © 2025 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests S.Z. is a founder and shareholder of Genemo, Inc. and Neurospan, LLC.
Figures

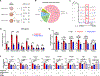

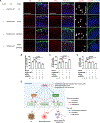

References
-
- Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, DeStafano AL, Bis JC, Beecham GW, Grenier-Boley B, et al. (2013). Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat. Genet 45, 1452–1458. 10.1038/ng.2802. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

