First-Tier Versus Last-Tier Trio Whole-Genome Sequencing for the Diagnosis of Pediatric-Onset Rare Diseases
- PMID: 40274276
- PMCID: PMC12405173
- DOI: 10.1111/cge.14760
First-Tier Versus Last-Tier Trio Whole-Genome Sequencing for the Diagnosis of Pediatric-Onset Rare Diseases
Abstract
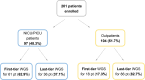
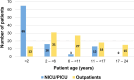
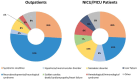
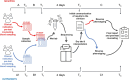
Despite advances in diagnostics, children with rare genetic disorders still face extended diagnostic odysseys, delaying appropriate clinical management, and placing burdens on families and healthcare resources. Whole-genome sequencing (WGS) offers a more comprehensive interrogation of the genome than other genetic tests, but its use in clinical practice remains limited. This study compared diagnostic rates, turnaround times, and clinical utility of first-tier versus last-tier trio-WGS for patients with suspected genetic pediatric-onset conditions, including 97 critical and 104 non-critical patients. Eighty-five patients (42.3%), including 57 (58.8%) critical and 28 (26.9%) non-critical patients, received a molecular diagnosis. The diagnostic rate was higher for first-tier (57%) than for last-tier (32.8%) trio-WGS. Of 121 causative variants identified, 19.8% would have been missed by whole-exome sequencing. Laboratory processing time was 4 days for all patients. The clinical setting had the greatest impact on time to reporting, averaging 5 days for critical patients versus 74 days for outpatients. WGS results impacted clinical decision-making for 34% of all critical and 14.3% of WGS-positive non-critical patients. This is the first Italian clinical study to demonstrate the diagnostic and clinical utility of a genome-first approach for both critical and non-critical patients with suspected genetic pediatric-onset disorders and feasibility in a public healthcare system.
Keywords: critical illness; delayed diagnosis; neonatal intensive care units; newborn infant diseases; outpatients; pediatric intensive care units; public health practice; rare diseases; whole genome sequencing.
© 2025 The Author(s). Clinical Genetics published by John Wiley & Sons Ltd.
Conflict of interest statement
Maria Iascone reports receiving TruSight Software Suite access, technical and editorial support, and open access publication fees through a collaboration agreement between Medical Genetics Laboratory of ASST Papa Giovanni XXIII and Illumina Inc. Daniela Piazzolla is an employee and shareholder of Illumina Inc.
Figures
References
-
- Shire , “Rare Disease Impact Report: Insights From Patients and the Medical Community,” 2013, https://globalgenes.org/wp‐content/uploads/2013/04/ShireReport‐1.pdf.
-
- Rosina E., Pezzani L., Apuril E., et al., “Comparison of First‐Tier Whole‐Exome Sequencing With a Multi‐Step Traditional Approach for Diagnosing Paediatric Outpatients: An Italian Prospective Study,” Molecular Genetics & Genomic Medicine 12, no. 1 (2024): e2316, 10.1002/mgg3.2316. - DOI - PMC - PubMed
MeSH terms
Grants and funding
- PNC0000003/National Plan for NRRP Complementary Investments (PNC, established with the decree-law 6 May 2021, n. 59, converted by law n. 101 of 2021) in the call for the funding of research initiatives for technologies and innovative trajectories in the health and care sectors (Directorial Decree n. 931 of 06-06-2022)
- "PG23/FROM 2017 Call for Independent Research" as part of the RARE - Rapid Analysis for Rapid carE - project
- Illumina Inc
- GSP21003/Telethon Foundation
LinkOut - more resources
Full Text Sources
Medical

