Serum analytes as predictors of disease recurrence and the duration of invasive disease-free survival in patients with triple negative breast cancer enrolled in the OXEL trial treated with immunotherapy, chemotherapy, or chemoimmunotherapy
- PMID: 40274284
- PMCID: PMC12020768
- DOI: 10.1136/jitc-2024-011379
Serum analytes as predictors of disease recurrence and the duration of invasive disease-free survival in patients with triple negative breast cancer enrolled in the OXEL trial treated with immunotherapy, chemotherapy, or chemoimmunotherapy
Abstract
Background: The OXEL study (NCT03487666) was a phase II trial of patients with triple negative breast cancer (TNBC) with residual disease following neoadjuvant chemotherapy, randomized to receive immunotherapy (anti-programmed cell death protein 1, nivolumab), chemotherapy (capecitabine), or chemoimmunotherapy. We previously reported on the primary endpoint of the OXEL trial, demonstrating that a peripheral immunoscore based on circulating immune cells reflecting immune activation was increased in patients treated with immunotherapy. However, compared with cell-based immune assays, sera assays are more cost-effective, less labor-intensive, and samples easier to obtain. Here, we report on differences in serum analytes between treatment arms and associations with clinical response.
Methods: Patients (n=38) were assayed for 97 serum analytes before and after 6 and 12 weeks of therapy. Serum analytes were assessed for changes with therapy, and as predictors of disease recurrence and the duration of invasive disease-free survival (iDFS) in both single analyte analyses and machine learning models.
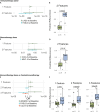
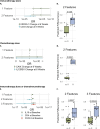
Results: Levels of specific analytes at baseline and changes in levels at early time points on treatment preceding recurrence were associated with eventual development of disease recurrence and/or the duration of iDFS. These associations varied based on the therapy patients received. Immunotherapy led to enrichment in pro-inflammatory analytes following treatment, whereas chemotherapy resulted in overall decreases. Changes seen in patients receiving chemoimmunotherapy more closely resembled those observed in patients receiving immunotherapy alone as opposed to chemotherapy alone. Furthermore, logistic regression and Cox proportional hazard models, developed using machine learning methods, demonstrated that combinations of serum analytes were more predictive of disease recurrence and iDFS duration than analyses of single serum analytes. Notably, the multivariable models that predicted patient outcomes were highly specific to the class of treatment patients received.
Conclusions: In patients with TNBC with residual disease after neoadjuvant chemotherapy, treatment with immunotherapy alone or chemoimmunotherapy resulted in enhanced immune activation compared with chemotherapy alone as measured by changes in serum analyte levels. Distinct serum analytes, both at baseline and as changes after therapy, predicted clinical outcomes for patients receiving immunotherapy alone, chemotherapy alone, or chemoimmunotherapy.
Trial registration number: NCT03487666.
Keywords: Biomarker; Breast Cancer; Chemotherapy; Immunotherapy.
© Author(s) (or their employer(s)) 2025. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ Group.
Conflict of interest statement
Competing interests: FL reports consulting for AstraZeneca, Pfizer, Eli-Lilly, Daiichi Sankyo; and institutional research funding from Merck, AstraZeneca, Zentalis, Ideaya, and Gilead. CI reports consultancies for AstraZeneca, Genentech, Gilead, ION, Merck, Medscape, MJH Holdings, Novartis, Pfizer, PUMA, and Seagen; Royalties from Wolters Kluwer (up-to-date) and McGraw Hill (Goodman and Gillman); and institutional research support from Tesaro/GSK, Seattle Genetics, Pfizer, AstraZeneca, BMS, Genentech, Novartis, and Regeneron. All other authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Medical
Research Materials
