STING aggravates ferroptosis-dependent myocardial ischemia-reperfusion injury by targeting GPX4 for autophagic degradation
- PMID: 40274801
- PMCID: PMC12022026
- DOI: 10.1038/s41392-025-02216-9
STING aggravates ferroptosis-dependent myocardial ischemia-reperfusion injury by targeting GPX4 for autophagic degradation
Abstract
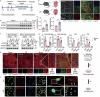
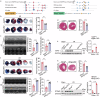
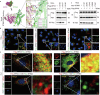
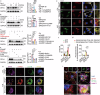
Despite advancements in interventional coronary reperfusion technologies following myocardial infarction, a notable portion of patients continue to experience elevated mortality rates as a result of myocardial ischemia-reperfusion (MI/R) injury. An in-depth understanding of the mechanisms underlying MI/R injury is crucial for devising strategies to minimize myocardial damage and enhance patient survival. Here, it is discovered that during MI/R, double-stranded DNA (dsDNA)-cyclic GMP-AMP synthase (cGAS)-stimulator of interferon genes (STING) signal accumulates, accompanied by high rates of myocardial ferroptosis. The specific deletion of cgas or Sting in cardiomyocytes, resulting in the inhibition of oxidative stress, has been shown to mitigate ferroptosis and I/R injury. Conversely, activation of STING exacerbates ferroptosis and I/R injury. Mechanistically, STING directly targets glutathione peroxidase 4 (GPX4) to facilitate its degradation through autophagy, by promoting the fusion of autophagosomes and lysosomes. This STING-GPX4 axis contributes to cardiomyocyte ferroptosis and forms a positive feedback circuit. Blocking the STING-GPX4 interaction through mutations in T267 of STING or N146 of GPX4 stabilizes GPX4. Therapeutically, AAV-mediated GPX4 administration alleviates ferroptosis induced by STING, resulting in enhanced cardiac functional recovery from MI/R injury. Additionally, the inhibition of STING by H-151 stabilizes GPX4 to reverse GPX4-induced ferroptosis and alleviate MI/R injury. Collectively, a novel autophagy-dependent ferroptosis mechanism is identified in this study. Specifically, STING autophagy induced by anoxia or ischemia-reperfusion leads to GPX4 degradation, thereby presenting a promising therapeutic target for heart diseases associated with I/R.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures




References
MeSH terms
Substances
Grants and funding
- 81970373-Zhang M/National Natural Science Foundation of China (National Science Foundation of China)
- 81920108003-Zhang C/National Natural Science Foundation of China (National Science Foundation of China)
- 82241203-Zhang C/National Natural Science Foundation of China (National Science Foundation of China)
- 82030051-Zhang Y/National Natural Science Foundation of China (National Science Foundation of China)
- 82200507/National Natural Science Foundation of China (National Science Foundation of China)
LinkOut - more resources
Full Text Sources
Research Materials

