Coxiella burnetii manipulates the lysosomal protease cathepsin B to facilitate intracellular success
- PMID: 40274809
- PMCID: PMC12022341
- DOI: 10.1038/s41467-025-59283-3
Coxiella burnetii manipulates the lysosomal protease cathepsin B to facilitate intracellular success
Abstract
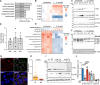
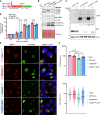
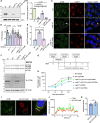
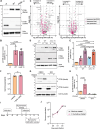
The obligate intracellular bacterium Coxiella burnetii establishes an intracellular replicative niche termed the Coxiella-containing vacuole (CCV), which has been characterised as a bacterially modified phagolysosome. How C. burnetii withstands the acidic and degradative properties of this compartment is not well understood. We demonstrate that the key lysosomal protease cathepsin B is actively and selectively removed from C. burnetii-infected cells through a mechanism involving the Dot/Icm type IV-B secretion system effector CvpB. Overexpression of cathepsin B leads to defects in CCV biogenesis and bacterial replication, indicating that removal of this protein represents a strategy to reduce the hostility of the intracellular niche. In addition, we show that C. burnetii infection of mammalian cells induces the secretion of a wider cohort of lysosomal proteins, including cathepsin B, to the extracellular milieu via a mechanism dependent on retrograde traffic. This study reveals that C. burnetii is actively modulating the hydrolase cohort of its replicative niche to promote intracellular success and demonstrates that infection incites the secretory pathway to maintain lysosomal homoeostasis.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures

References
-
- Bird, L. E., Edgington-Mitchell, L. E. & Newton, H. J. Eat, prey, love: pathogen-mediated subversion of lysosomal biology. Curr. Opin. Immun.83, 102344 (2023). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

