Defective autophagy in CD4 T cells drives liver fibrosis via type 3 inflammation
- PMID: 40274816
- PMCID: PMC12022296
- DOI: 10.1038/s41467-025-59218-y
Defective autophagy in CD4 T cells drives liver fibrosis via type 3 inflammation
Abstract
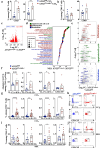
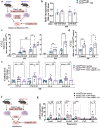
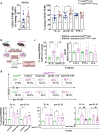
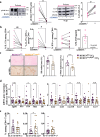
Conventional CD4 T cells represent a major source of inflammatory mediators that drive progression of chronic liver disease to fibrosis and to end-stage cirrhosis. Identification of T cell pathways that limits the inflammatory response could thus have therapeutic relevance. Here we show, using both human samples and mouse models, that autophagy is deficient in CD4 T cells from patients with advanced fibrosis, and that loss of autophagy following genomic deletion of ATG5 in T cells is associated with the emergence of pathogenic IL-17A + IFN-γ + Th17 T cells that drive liver fibrosis in mice. Mechanistically, liver CD4 T cells lacking autophagy display a Th17 glycolytic phenotype associated with enhanced type 3 cytokine (i.e., IL-17A and GM-CSF) release, shifting hepatic myofibroblasts, hepatocytes and macrophages toward a proinflammatory phenotype. We also show that autophagy can be rescued in CD4 T cells from patients with extensive liver fibrosis, leading to decreased frequency of pathogenic Th17 cells and reduced GM-CSF levels; in addition, limited fibrosis is observed in mice in which Rubicon, a negative regulator of autophagy, is deleted specifically in their T cells. Our findings thus implicate autophagy in CD4 T cells as a key therapeutic target to control inflammation-driven fibrosis during chronic liver injury.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures


References
-
- Gilgenkrantz, H., Sayegh, R. A. & Lotersztajn, S. Immunoregulation of liver fibrosis: new opportunities for antifibrotic therapy. Annu Rev. Pharm. Toxicol.65, 281–299 (2025). - PubMed
-
- Mallat, A. & Lotersztajn, S. Cellular mechanisms of tissue fibrosis. 5. novel insights into liver fibrosis. Am. J. Physiol. Cell Physiol.305, C789–C799 (2013). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

