Age-Dependent Regulation of Hippocampal Inflammation by the Mitochondrial Translocator Protein in Mice
- PMID: 40275629
- PMCID: PMC12151901
- DOI: 10.1111/acel.70039
Age-Dependent Regulation of Hippocampal Inflammation by the Mitochondrial Translocator Protein in Mice
Abstract
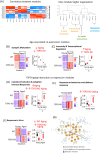
The mitochondrial translocator protein (TSPO) is a biomarker of inflammation associated with neurodegenerative diseases, widely regarded to be upregulated in the aging brain. Here we investigated the interaction between aging and TSPO immunomodulatory function in the mouse hippocampus, a region severely affected in Alzheimer's Disease (AD). Surprisingly, we found that TSPO levels were decreased in brain innate immune populations in aging. Aging resulted in a reversal of TSPO knockout transcriptional signatures following inflammatory insult. TSPO deletion drastically exacerbated inflammatory transcriptional responses in the aging hippocampus, while dampening inflammation in the young hippocampus. This age-dependent effect of TSPO was linked to NF-kβ and interferon regulatory transcriptional networks. Drugs that disrupt the cell cycle and induce DNA damage, such as heat shock protein and topoisomerase inhibitors, were identified to mimic the inflammatory transcriptional signature characterizing aging in TSPO knockout mice most closely. These findings indicate that TSPO plays a protective role in brain aging. This TSPO-aging interaction is an important consideration in the interpretation of TSPO-targeted biomarker and therapeutic studies, as well as in vitro studies that cannot model the aging brain.
Keywords: LPS; aging; hippocampus; mitochondria; neuroinflammation; translocator protein.
© 2025 The Author(s). Aging Cell published by Anatomical Society and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures





References
-
- Ang, H. P. , Makpol S., Nasaruddin M. L., et al. 2023. “Lipopolysaccharide‐Induced Delirium‐Like Behaviour in a Rat Model of Chronic Cerebral Hypoperfusion Is Associated With Increased Indoleamine 2,3‐Dioxygenase Expression and Endotoxin Tolerance.” International Journal of Molecular Sciences 24, no. 15: 12248. 10.3390/ijms241512248. - DOI - PMC - PubMed
-
- Bahaidrah, K. A. , Alzahrani N. A., Aldhahri R. S., Mansouri R. A., and Alghamdi B. S.. 2022. “Effects of Different Lipopolysaccharide Doses on Short‐ and Long‐Term Spatial Memory and Hippocampus Morphology in an Experimental Alzheimer's Disease Model.” Clinical and Translational Neuroscience 6, no. 3: 20.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

