Natural variation in potassium deficiency responses among Arabidopsis thaliana accessions
- PMID: 40275769
- PMCID: PMC12290283
- DOI: 10.1093/pcp/pcaf041
Natural variation in potassium deficiency responses among Arabidopsis thaliana accessions
Abstract
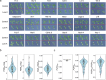
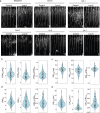
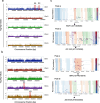
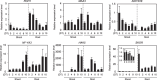
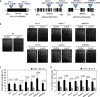
Potassium (K) is a key nutrient essential for plant growth, and its deficiency induces various adaptative responses in plants; however, the mechanisms underlying these responses remain unclear. In the present study, we explored the natural variation in K deficiency responses among 100 naturally occurring accessions of Arabidopsis thaliana and then performed a genome-wide association study (GWAS) to identify the genetic loci associated with these responses. All 100 Arabidopsis accessions showed significant differences in several traits under K deficiency, including shoot and root growth, photosynthetic activity, and inorganic ion contents. The results indicated that the reduction in K+ content due to K deficiency was correlated more significantly with decreases in the number and total length of lateral roots than with decreases in primary root length and shoot growth. Furthermore, GWAS and subsequent analyses of relevant mutants and transgenic plants suggested that several genes, which have not yet been shown to play a role in the K deficiency response, are associated with the number and/or total length of lateral roots under K deficiency. The identified genes are ROH1, NF-YA3, MAA3, and AtDTX28, which encode an exocyst subunit EXO70A1 interacting protein, an NF-Y type transcription factor, a female gametophyte development-related protein, and a MATE efflux family protein, respectively. These findings provide new insights into the mechanisms underlying K deficiency responses.
Keywords: Arabidopsis thaliana; GWAS; lateral root development; natural variation; potassium deficiency.
© The Author(s) 2025. Published by Oxford University Press on behalf of Japanese Society of Plant Physiologists.
Conflict of interest statement
The authors have no conflicts of interest to declare.
Figures



References
-
- Alemán F., Nieves-Cordones M., Martínez V. and Rubio F. (2009) Differential regulation of the HAK5 genes encoding the high-affinity K+ transporters of Thellungiella halophila and Arabidopsis thaliana. Environ. Exp. Bot. 65: 263–269. doi: 10.1016/j.envexpbot.2008.09.011 - DOI
-
- Amtmann A., Hammond J.P., Armengaud P. and White P.J. (2006) Nutrient sensing and signalling in plants: potassium and phosphorus. Adv. Bot. Res. 43: 209–257.
-
- Armengaud P., Sulpice R., Mille A.J., Stitt M., Amtmann A. and Gibon Y. (2009) Multilevel analysis of primary metabolism provides new insights into the role of potassium nutrition for glycolysis and nitrogen assimilation in Arabidopsis roots. Plant Physiol. 150: 772–785. doi: 10.1104/pp.108.133629 - DOI - PMC - PubMed
-
- Bell M.J., Ransom M.D., Thompson M.L., Hinsinger P., Florence A.M., Moody P.W., et al. (2021) Considering soil potassium pools with dissimilar plant availability. Improving Potassium Recommendations for Agricultural Crops. Edited by Murrell, T.S., Mikkelsen, R.L., Sulewski, G., Norton, R. and Thompson, M.L. pp. 163–190. Springer International Publishing AG, Cham.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

