Origin and development of uniparental and polyploid blastomeres
- PMID: 40276758
- PMCID: PMC12020880
- DOI: 10.1016/j.isci.2025.112337
Origin and development of uniparental and polyploid blastomeres
Abstract
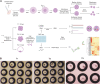
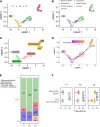
Whole-genome (WG) abnormalities, such as uniparental diploidy and triploidy, cause fetal death. Occasionally, they coexist with biparental diploid cells in live births. Understanding the origin and early development of WG abnormal blastomeres is crucial for explaining the formation of androgenotes, gynogenotes, triploidy, chimerism, and mixoploidy. By haplotyping 118 bovine blastomeres from the first cleavages, we identified that heterogoneic division occurs in both multipolar and bipolar cleaving zygotes. During heterogoneic division, parental genomes segregate into distinct blastomeres, resulting in the coexistence of uniparental and biparental diploid or polyploid cells. After culturing the totipotent blastomeres to three preimplantation stages and exploring transcriptomes of 446 cells, we discovered that stress responses contribute to developmental impairment in WG abnormal cells, resulting in either cell arrest or blastocyst formation. Their dominance in preimplantation embryos represents an overlooked cause of abnormal development. Haplotype-based screening could improve in vitro fertilization outcomes.
Keywords: Molecular biology; Omics.
© 2025 Published by Elsevier Inc.
Conflict of interest statement
T.V. and J.R.V. are co-inventors on licensed patents WO/2011/157846 (Methods for haplotyping single cells), WO/2014/053664 (High-throughput genotyping by sequencing low amounts of genetic material), and WO/2015/028576 (Haplotyping and copy number typing using polymorphic variant allelic frequencies).
Figures





References
-
- Zaragoza M.V., Surti U., Redline R.W., Millie E., Chakravarti A., Hassold T.J. Parental origin and phenotype of triploidy in spontaneous abortions: Predominance of diandry and association with the partial hydatidiform mole. Am. J. Hum. Genet. 2000;66:1807–1820. doi: 10.1086/302951. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

