Downregulation of rRNA synthesis by BCL-2 induces chemoresistance in diffuse large B cell lymphoma
- PMID: 40276769
- PMCID: PMC12020883
- DOI: 10.1016/j.isci.2025.112333
Downregulation of rRNA synthesis by BCL-2 induces chemoresistance in diffuse large B cell lymphoma
Abstract
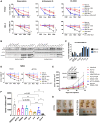
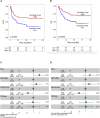
Overexpression of the antiapoptotic oncogene BCL-2 predicts poor prognosis in diffuse large B cell lymphoma (DLBCL) treated with anthracycline-based chemoimmunotherapy. Anthracyclines exert antitumor effects by multiple mechanisms including inhibition of ribosome biogenesis (RiBi) through rRNA synthesis blockade. RiBi inhibitors induce p53 stabilization through the ribosomal proteins-MDM2-p53 pathway, with stabilized p53 levels depending on baseline rRNA synthesis rate. We found that the BH3-mimetic venetoclax could not fully reverse BCL-2-mediated resistance to RiBi inhibitors in DLBCL cells. BCL-2 overexpression was associated with decreased baseline rRNA synthesis rate, attenuating p53 stabilization by RiBi inhibitors. Drugs stabilizing p53 irrespective of RiBi inhibition reversed BCL-2-induced resistance in vitro and in vivo, restoring p53 activation and apoptosis. A small nucleolar size, indicative of low baseline rRNA synthesis, correlated with high BCL-2 levels and poor outcomes in DLBCL patients. These findings uncover alternative BCL-2-dependent chemoresistance mechanisms, providing a rationale for specific combination strategies in BCL-2 positive lymphomas.
Keywords: Cancer; Cell biology.
© 2025 The Author(s).
Conflict of interest statement
E.D., research funding: Takeda, ADC-Therapeutics, and Incyte; speaker’s bureau: Roche, Incyte, and Abbvie; advisory board: Astra Zeneca, Lilly, Abbvie, Roche, Gilead, Takeda, and Sobi. F.B., research support: Roche and Menarini; speaker’s bureau: Pfizer. P.L.Z., consultant: MSD, Eusapharma, and Novartis; speaker’s bureau: Celltrion, Gilead, Jassen-Cilag, BMS, Servier, MSD, Astrazeneca, Tekada, Roche, Eusapharma, Kyowa Kirin, Novartis, Incyte, and BeiGene; advisory board: Secura Bio, Celltrion, Gilead, Jassen-Cilag, BMS, Servier, Sandoz, MSD, Astrazeneca, Tekada, Roche, Eusapharma, Kyowa Kirin, Novartis, ADC Terap., Incyte, and BeiGene. S.P., speaker’s bureau: Lilly, Takeda, BeiGene, Stemline, and Roche; advisory board: Lilly, Stemline, and Diatech.
Figures

References
-
- Crump M., Neelapu S.S., Farooq U., Van Den Neste E., Kuruvilla J., Westin J., Link B.K., Hay A., Cerhan J.R., Zhu L., et al. Outcomes in refractory diffuse large B-cell lymphoma: Results from the international SCHOLAR-1 study. Blood. 2017;130:1800–1808. doi: 10.1182/blood-2017-03-769620. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

