KIF5A p.Pro986Leu Risk Variant and Accelerated Progression of Amyotrophic Lateral Sclerosis
- PMID: 40276954
- PMCID: PMC12257138
- DOI: 10.1002/acn3.70059
KIF5A p.Pro986Leu Risk Variant and Accelerated Progression of Amyotrophic Lateral Sclerosis
Abstract
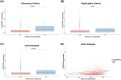
This study explored the impact of KIF5A rs113247976 (p.Pro986Leu), a risk allele for amyotrophic lateral sclerosis (ALS), on phenotypic variability in two Italian ALS cohorts (discovery, n = 865; replication, n = 1174). The minor allele (T) frequency was 0.015. No patients were homozygous (TT), allowing comparison between wild type and heterozygous carriers only. Heterozygous carriers showed faster disease progression (ALSFRS-R preslope). Findings were validated across both cohorts. Multiple linear regression identified p.Leu986 and age at onset as ALSFRS-R preslope predictors. In conclusion, heterozygous p.Leu986 in KIF5A is associated with faster ALS progression, supporting its consideration for genetic screening in clinical trials.
Keywords: KIF5A; alleles; amyotrophic lateral sclerosis; disease progression; motor neurons.
© 2025 The Author(s). Annals of Clinical and Translational Neurology published by Wiley Periodicals LLC on behalf of American Neurological Association.
Conflict of interest statement
V.S. received compensation for consulting services and/or speaking activities from AveXis, Cytokinetics, Italfarmaco, Liquidweb Srl, Novartis Pharma AG, and Zambon Biotech SA. He is on the Editorial Board of Amyotrophic Lateral Sclerosis and Frontotemporal Degeneration, European Neurology, American Journal of Neurodegenerative Diseases, Frontiers in Neurology, and Exploration of Neuroprotective Therapy. N.T. received compensation for consulting services from Amylyx Pharmaceuticals, Biogen, Italfarmaco, and Zambon Biotech SA. He is Associate Editor for Frontiers in Aging Neuroscience.
Figures
References
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
