Dietary supplementation of male mice with inorganic, organic or nanoparticle selenium preparations: evidence supporting a putative gut-thyroid-male fertility axis
- PMID: 40277453
- PMCID: PMC12035940
- DOI: 10.1080/13510002.2025.2495367
Dietary supplementation of male mice with inorganic, organic or nanoparticle selenium preparations: evidence supporting a putative gut-thyroid-male fertility axis
Abstract
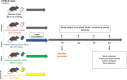
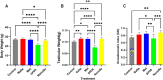
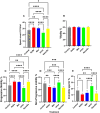
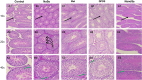
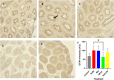
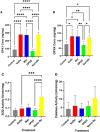
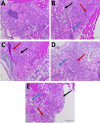
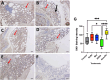
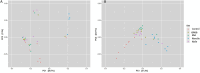
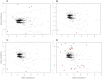
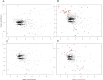
Selenium (Se) is linked to physiological homeostasis. Male mice (n = 8/group) were fed control (AIN93G) or diets enriched in sodium selenite (NaSe, 5.6 ppm), methylselenocysteine (Met, 4.7 ppm), diphenyl diselenide (DPDS, 14.2 ppm), or nanoselenium (NanoSe, 2.7 ppm); dietary Se ascertained by inductively-coupled plasma mass spectrometry. At 4 weeks testes, sperm, thyroids, blood and stool were collected to assess histoarchitecture, circulating hormones (thyroxine, T4; triiodothyronine, T3; thyroid stimulating hormone, TSH) and gut microbiome (16S rRNAV3-V4 amplicon sequencing). Supplemented NaSe, Met, and NanoSe increased plasma testosterone and testis glutathione peroxidases (GPx-1/4) while testicular superoxide dismutase and catalase increased slightly in the NanoSe group indicating a selective antioxidant response. Overall, NanoSe and NaSe enhanced male reproductive factors. All thyroids isolated from Se-supplemented mice contained marginal vacuoles and a lower follicle area vs control. Nano-Se enhanced thyroidiodothyronine deiodinase-1 (DIO1) expression however, thyroid GPx-1/4 remained unchanged. Supplemented NaSe and DPDSl increased plasma T3/T4 ratio, while plasma TSH was unchanged. Microbiome analyses showed that NanoSe was most efficacious in altering composition (judged by α-diversity, Shannon index and taxon richness) while the NaSe diet showed the greatest overall change in α-diversity. Dietary Se supplementation, particularly encapsulated NanoSe, may improve male fertility factors by enhancing the gut-thyroid-fertility axis.
Keywords: Dietary selenium; male fertility; microbiome; microbiota; nanoselenium; testosterone; thyroid; thyroid hormone.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures





References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous
