Strategies for p53 Activation and Targeted Inhibitors of the p53-Mdm2/MdmX Interaction
- PMID: 40277907
- PMCID: PMC12025665
- DOI: 10.3390/cells14080583
Strategies for p53 Activation and Targeted Inhibitors of the p53-Mdm2/MdmX Interaction
Abstract
p53 is a tumor suppressor gene and is regarded as one of the most crucial genes in protecting humans against cancer. The protein Mdm2 and its homolog MdmX serve as negative regulators of p53. In nearly half of cancer cells, there is an overexpression of Mdm2 and MdmX, which inhibit p53 activity. Furthermore, Mdm2's E3 ubiquitin ligase activity promotes the ubiquitination and degradation of p53. Therefore, blocking the interaction between p53 and Mdm2/MdmX to prevent the degradation of wild-type p53 is an effective strategy for inhibiting tumor growth. This paper primarily discusses the regulatory relationship between p53, MdmX and Mdm2, and provides a review of the current status of p53-Mdm2/MdmX inhibitors. It aims to offer a theoretical foundation and research direction for the future discovery and design of targeted inhibitors against the p53-Mdm2/MdmX interaction.
Keywords: Mdm2; MdmX; cancer; inhibitors; p53.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



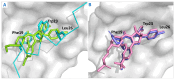
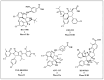

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

