Integrative Analysis of DNA Methylation and microRNA Reveals GNPDA1 and SLC25A16 Related to Biopsychosocial Factors Among Taiwanese Women with a Family History of Breast Cancer
- PMID: 40278313
- PMCID: PMC12028518
- DOI: 10.3390/jpm15040134
Integrative Analysis of DNA Methylation and microRNA Reveals GNPDA1 and SLC25A16 Related to Biopsychosocial Factors Among Taiwanese Women with a Family History of Breast Cancer
Abstract
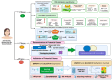
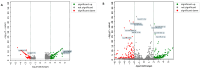
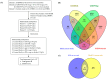
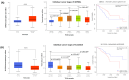
Biopsychosocial factors, including family history, influence the development of breast cancer. Malignancies in women with a family history of breast cancer may be detectable based on DNA methylation and microRNA. Objectives: The present study extended an integrative analysis of DNA methylation and microRNA to identify genes associated with biopsychosocial factors. Methods: We identified 3060 healthy women from the Taiwan Biobank and included 32 blood plasma samples for analysis of biopsychosocial factors and epigenetic changes. GEO databases and bioinformatics approaches were used for the identification and validation of potential genes. Results: Our integrative analysis revealed GNPDA1 and SLC25A16 as potential genes. Age, a family history of cancer, and alcohol consumption were associated with GNPDA1 and SLC25A16 based on the current data set and the GEO data set. GNPDA1 and SLC25A16 exhibited significant expression in breast cancer tissues based on UALCAN analysis, where they were overexpressed and underexpressed, respectively. Through a MethSurv analysis, GNPDA1 hypomethylation and SLC25A16 hypermethylation were associated with poor prognoses in terms of overall survival in breast cancer. Moreover, through a MetaCore functional enrichment analysis, GNPDA1 and SLC25A16 were associated with the BRCA1, BRCA2, and pro-oncogenic actions of the androgen receptor in breast cancer. Further, GNPDA1 and SLC25A16 were enriched in known targets of approved cancer drugs as potential genes associated with breast cancer. Conclusions: These two genes might serve as biomarkers for the early detection of breast cancer, especially for women with a family history of breast cancer.
Keywords: DNA methylation; biopsychosocial factors; family history of breast cancer; integrative analysis; microRNA.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

Similar articles
-
Glucosamine-6-Phosphate Isomerase 1 Promotes Tumor Progression and Indicates Poor Prognosis in Hepatocellular Carcinoma.Cancer Manag Res. 2020 Jun 24;12:4923-4935. doi: 10.2147/CMAR.S250094. eCollection 2020. Cancer Manag Res. 2020. PMID: 32606980 Free PMC article.
-
Familial and racial determinants of tumour suppressor genes promoter hypermethylation in breast tissues from healthy women.J Cell Mol Med. 2010 Jun;14(6B):1468-75. doi: 10.1111/j.1582-4934.2009.00924.x. Epub 2009 Oct 3. J Cell Mol Med. 2010. PMID: 19799643 Free PMC article.
-
LAT, HOXD3 and NFE2L3 identified as novel DNA methylation-driven genes and prognostic markers in human clear cell renal cell carcinoma by integrative bioinformatics approaches.J Cancer. 2019 Oct 22;10(26):6726-6737. doi: 10.7150/jca.35641. eCollection 2019. J Cancer. 2019. PMID: 31777602 Free PMC article.
-
Potential Prognostic Biomarkers of NIMA (Never in Mitosis, Gene A)-Related Kinase (NEK) Family Members in Breast Cancer.J Pers Med. 2021 Oct 26;11(11):1089. doi: 10.3390/jpm11111089. J Pers Med. 2021. PMID: 34834441 Free PMC article.
-
Hypermethylation of TMEM240 predicts poor hormone therapy response and disease progression in breast cancer.Mol Med. 2022 Jun 17;28(1):67. doi: 10.1186/s10020-022-00474-9. Mol Med. 2022. PMID: 35715741 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

