Specific Aspects of SELEX Protocol: Different Approaches for ssDNA Generation
- PMID: 40278510
- PMCID: PMC12029403
- DOI: 10.3390/mps8020036
Specific Aspects of SELEX Protocol: Different Approaches for ssDNA Generation
Abstract
Background: Synthetic DNA aptamers are a class of molecules with potential applications in medicine, serving as molecular sensors or ligands for targeted drug delivery. Systematic evolution of ligands by exponential enrichment (SELEX) is a technology for selecting functional aptamers that was first reported three decades ago and has been actively developed since. SELEX involves multiple iterations of two fundamental steps: (i) target affinity-based partitioning of aptamers from a random library and (ii) amplification of selected aptamers by PCR, followed by isolation of single-stranded DNA (ssDNA). SELEX protocols have diversified considerably, with numerous variations possible for each step. This heterogeneity makes it challenging to identify optimal methods. Comparative analysis of different approaches for the major stages of SELEX is therefore of considerable practical importance.
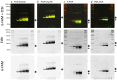
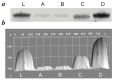
Methods: Four widely used methods for ssDNA generation were performed in parallel: (a) PCR followed by digestion of the antisense strand with exonuclease lambda, (b) PCR with an extended primer followed by size-dependent strand separation using denaturing PAGE, (c) asymmetric PCR, and (d) asymmetric PCR with a primer-blocker.
Results: The specificity, efficiency, reproducibility, and duration of each method were compared.
Conclusions: Asymmetric PCR with a primer-blocker yielded the most favorable results.
Keywords: PCR; SELEX; aptamer; asymmetric PCR; single-stranded DNA.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures





Similar articles
-
Optimisation of denaturing ion pair reversed phase HPLC for the purification of ssDNA in SELEX.J Chromatogr A. 2024 Mar 29;1719:464699. doi: 10.1016/j.chroma.2024.464699. Epub 2024 Feb 2. J Chromatogr A. 2024. PMID: 38382212
-
Single-stranded DNA (ssDNA) production in DNA aptamer generation.Analyst. 2012 Mar 21;137(6):1307-15. doi: 10.1039/c2an15905h. Epub 2012 Feb 7. Analyst. 2012. PMID: 22314701 Review.
-
Challenges to design and develop of DNA aptamers for protein targets. I. Optimization of asymmetric PCR for generation of a single stranded DNA library.Iran J Pharm Res. 2014 Winter;13(Suppl):133-41. Iran J Pharm Res. 2014. PMID: 24711839 Free PMC article.
-
Upgrading SELEX technology by using lambda exonuclease digestion for single-stranded DNA generation.Molecules. 2009 Dec 24;15(1):1-11. doi: 10.3390/molecules15010001. Molecules. 2009. PMID: 20110867 Free PMC article.
-
SELEX: Critical factors and optimization strategies for successful aptamer selection.Biotechnol Appl Biochem. 2022 Oct;69(5):1771-1792. doi: 10.1002/bab.2244. Epub 2021 Sep 3. Biotechnol Appl Biochem. 2022. PMID: 34427974 Free PMC article. Review.
References
Grants and funding
LinkOut - more resources
Full Text Sources

