Building Pathogen Genomic Sequencing Capacity in Africa: Centre for Epidemic Response and Innovation Fellowship
- PMID: 40278763
- PMCID: PMC12030795
- DOI: 10.3390/tropicalmed10040090
Building Pathogen Genomic Sequencing Capacity in Africa: Centre for Epidemic Response and Innovation Fellowship
Abstract
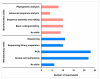
The World Health Organization African region has the greatest infectious disease burden in the world. However, many African countries have limited capacity to rapidly detect, report, and respond to public health events. The Centre for Epidemic Response and Innovation (CERI), KwaZulu-Natal Research Innovation and Sequencing Platform (KRISP) in South Africa, and global Climate Amplified Diseases and Epidemics (CLIMADE) consortium are investing in building the capacity of African scientists in pathogen genomics and bioinformatics. A two-week long (11-21 April 2023) intensive training in wet-laboratory genomic data production, bioinformatics, and phylogenetic analyses of viral and bacterial pathogens was held in Cape Town, South Africa. Training was provided to 36 fellows with diverse backgrounds from 16 countries, 14 of which were low- and middle-income African countries. In this report, we, the fellows, share our collective experiences and describe how the learnt skills have been integrated into the operations of our home institutions to advance genomic surveillance capabilities. We identified the in-person and hands-on learning format of the training, taught by genomics experts and field application specialists, as the most impactful elements of this training event. Adaptation and miniaturisation of protocols to detect other pathogens is a great enhancement over the traditional method of using a single protocol for a pathogen. We note the duration of the training as the largest limiting factor, particularly for the computationally intensive bioinformatics sessions. We recommend this programme continue to build pathogen genomics capacity in Africa.
Keywords: bioinformatics; capacity building; genomic surveillance; lessons; pathogens; training.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
-
- Chabuka L., Choga W.T., Mavian C.N., Moir M., Tegally H., Wilkinson E., Naidoo Y., Inward R., Morgenstern C., Bhatt S., et al. Genomic Epidemiology of the Cholera Outbreak in Malawi 2022–2023. medRxiv. 2023 doi: 10.1101/2023.08.22.23294324. medRxiv:2023.08.22.23294324. - DOI
-
- Jackson B.R., Tarr C., Strain E., Jackson K.A., Conrad A., Carleton H., Katz L.S., Stroika S., Gould L.H., Mody R.K., et al. Implementation of Nationwide Real-Time Whole-Genome Sequencing to Enhance Listeriosis Outbreak Detection and Investigation. Clin. Infect. Dis. 2016;63:380–386. doi: 10.1093/cid/ciw242. - DOI - PMC - PubMed
-
- Oude Munnink B.B., Nieuwenhuijse D.F., Stein M., O’Toole Á., Haverkate M., Mollers M., Kamga S.K., Schapendonk C., Pronk M., Lexmond P., et al. Rapid SARS-CoV-2 Whole-Genome Sequencing and Analysis for Informed Public Health Decision-Making in the Netherlands. Nat. Med. 2020;26:1405–1410. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

