Noninvasive prognostication of hepatocellular carcinoma based on cell-free DNA methylation
- PMID: 40279344
- PMCID: PMC12026916
- DOI: 10.1371/journal.pone.0321736
Noninvasive prognostication of hepatocellular carcinoma based on cell-free DNA methylation
Abstract
Background: The current noninvasive prognostic evaluation methods for hepatocellular carcinoma (HCC), which are largely reliant on radiographic imaging features and serum biomarkers such as alpha-fetoprotein (AFP), have limited effectiveness in discriminating patient outcomes. Identification of new prognostic biomarkers is a critical unmet need to improve treatment decision-making. Epigenetic changes in cell-free DNA (cfDNA) have shown promise in early cancer diagnosis and prognosis. Thus, we aim to evaluate the potential of cfDNA methylation as a noninvasive predictor for prognostication in patients with active, radiographically viable HCC.
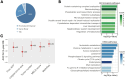
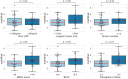
Methods: Using Illumina HumanMethylation450 array data of 377 HCC tumors and 50 adjacent normal tissues obtained from The Cancer Genome Atlas (TCGA), we identified 158 HCC-related DNA methylation markers associated with overall survival (OS). This signature was further validated in 29 HCC tumor tissue samples. Subsequently, we applied the signature to an independent cohort of 52 patients with plasma cfDNA samples by calculating the cfDNA methylation-based risk score (methRisk) via random survival forest models with 10-fold cross-validation for the prognostication of OS.
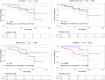
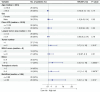
Results: The cfDNA-based methRisk showed strong discriminatory power when evaluated as a single predictor for OS (3-year AUC = 0.81, 95% CI: 0.68-0.94). Integrating the methRisk with existing risk indices like Barcelona clinic liver cancer (BCLC) staging significantly improved the noninvasive prognostic assessments for OS (3-year AUC = 0.91, 95% CI: 0.80-1), and methRisk remained an independent predictor of survival in the multivariate Cox model (P = 0.007).
Conclusions: Our study serves as a pilot study demonstrating that cfDNA methylation biomarkers assessed from a peripheral blood draw can stratify HCC patients into clinically meaningful risk groups. These findings indicate that cfDNA methylation is a promising noninvasive prognostic biomarker for HCC, providing a proof-of-concept for its potential clinical utility and laying the groundwork for broader applications.
Copyright: © 2025 Hu et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
We have read the journal’s policy and the authors of this manuscript have the following competing interests: W.L. and X.J.Z. are co-founders of EarlyDiagnostics. X.J.Z serves on the Board of Directors and has an executive leadership position at EarlyDiagnostics. W.L. and X.J.Z. are stockholders of EarlyDiagnostics. S.L., M.L.S., W.Z., and Y.Z. have stock options with EarlyDiagnostics. W.Z., W.L., Y.Z., and V.A. are consultants for EarlyDiagnostics. The other authors declare no competing interests.
Figures