MAX inactivation deregulates the MYC network and induces neuroendocrine neoplasia in multiple tissues
- PMID: 40279415
- PMCID: PMC12024646
- DOI: 10.1126/sciadv.adt3177
MAX inactivation deregulates the MYC network and induces neuroendocrine neoplasia in multiple tissues
Abstract
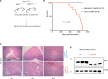
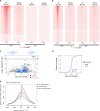
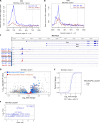
The MYC transcription factor requires MAX for DNA binding and widespread activation of gene expression in both normal and neoplastic cells. Inactivating mutations in MAX are associated with a subset of neuroendocrine cancers including pheochromocytoma, pituitary adenoma, and small cell lung cancer. Neither the extent nor the mechanisms of MAX tumor suppression are well understood. Deleting Max across multiple mouse neuroendocrine tissues, we find that Max inactivation alone produces pituitary adenomas, while Max inactivation cooperates with Rb1/Trp53 loss to accelerate medullary thyroid C cell and pituitary adenoma development. In the thyroid tumor cell lines, MAX loss triggers a marked shift in genomic occupancy by other members of the MYC network (MNT, MLX, MondoA) supporting metabolism, survival, and proliferation of neoplastic neuroendocrine cells. Our work reveals MAX as a broad suppressor of neuroendocrine tumorigenesis through its ability to maintain a balance of genomic occupancies among the diverse transcription factors in the MYC network.
Figures





Update of
-
MAX inactivation deregulates the MYC network and induces neuroendocrine neoplasia in multiple tissues.bioRxiv [Preprint]. 2024 Sep 24:2024.09.21.614255. doi: 10.1101/2024.09.21.614255. bioRxiv. 2024. Update in: Sci Adv. 2025 Apr 25;11(17):eadt3177. doi: 10.1126/sciadv.adt3177. PMID: 39386474 Free PMC article. Updated. Preprint.
References
-
- Blackwood E. M., Eisenman R. N., Max: A helix-loop-helix zipper protein that forms a sequence-specific DNA binding complex with Myc. Science 251, 1211–1217 (1991). - PubMed
-
- Amati B., Dalton S., Brooks M. W., Littlewood T. D., Evan G. I., Land H., Transcriptional activation by the human c-Myc oncoprotein in yeast requires interaction with Max. Nature 359, 423–426 (1992). - PubMed
-
- Ayer D. E., Kretzner L., Eisenman R. N., Mad: A heterodimeric partner for Max that antagonizes Myc transcriptional activity. Cell 72, 211–222 (1993). - PubMed
-
- Nair S. K., Burley S. K., X-ray structures of myc-max and mad-max recognizing DNA. Molecular bases of regulation by proto-oncogenic transcription factors. Cell 112, 193–205 (2003). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

