Prolonged airway explant culture enables study of health, disease, and viral pathogenesis
- PMID: 40279421
- PMCID: PMC12024639
- DOI: 10.1126/sciadv.adp0451
Prolonged airway explant culture enables study of health, disease, and viral pathogenesis
Abstract
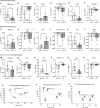
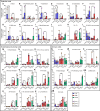
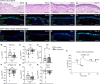
In vitro models play a major role in studying airway physiology and disease. However, the native lung's complex tissue architecture and nonepithelial cell lineages are not preserved in these models. Ex vivo tissue models could overcome in vitro limitations, but methods for long-term maintenance of ex vivo tissue have not been established. Here, we describe methods to culture human large airway explants, small airway explants, and precision-cut lung slices for at least 14 days. Human airway explants recapitulate genotype-specific electrophysiology; characteristic epithelial, endothelial, stromal, and immune cell populations; and model viral infection after 14 days in culture. These methods also maintain mouse, rabbit, and pig tracheal explants. Notably, intact airway tissue can be cryopreserved, thawed, and used to generate viable explants with recovery of function 14 days postthaw. These studies highlight the broad applications of airway tissue explants and their use as translational intermediates between in vitro and in vivo studies.
Figures





Update of
-
Prolonged airway explant culture enables study of health, disease, and viral pathogenesis.bioRxiv [Preprint]. 2024 Feb 5:2024.02.03.578756. doi: 10.1101/2024.02.03.578756. bioRxiv. 2024. Update in: Sci Adv. 2025 Apr 25;11(17):eadp0451. doi: 10.1126/sciadv.adp0451. PMID: 38370820 Free PMC article. Updated. Preprint.
References
-
- Fulcher M. L., Randell S. H., Human nasal and tracheo-bronchial respiratory epithelial cell culture. Methods Mol. Biol. 945, 109–121 (2013). - PubMed
-
- Randell S. H., Fulcher M. L., O’Neal W., Olsen J. C., Primary epithelial cell models for cystic fibrosis research. Methods Mol. Biol. 742, 285–310 (2011). - PubMed
-
- Merkle H., Ditzinger G., Lang S., Peter H., Schmidt M., In vitro cell models to study nasal mucosal permeability and metabolism. Adv. Drug Deliv. Rev. 29, 51–79 (1998). - PubMed
-
- Wadell C., Björk E., Camber O., Permeability of porcine nasal mucosa correlated with human nasal absorption. Eur. J. Pharm. Sci. 18, 47–53 (2003). - PubMed
-
- Chemuturi N. V., Hayden P., Klausner M., Donovan M. D., Comparison of human tracheal/bronchial epithelial cell culture and bovine nasal respiratory explants for nasal drug transport studies. J. Pharm. Sci. 94, 1976–1985 (2005). - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

