The mechanism of peptidoglycan O-acetylation in Gram-negative bacteria typifies bacterial MBOAT-SGNH acyltransferases
- PMID: 40280421
- PMCID: PMC12148441
- DOI: 10.1016/j.jbc.2025.108531
The mechanism of peptidoglycan O-acetylation in Gram-negative bacteria typifies bacterial MBOAT-SGNH acyltransferases
Abstract
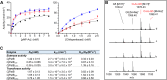
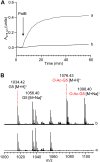
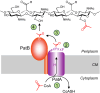
Bacterial cell envelope polymers are commonly modified with acyl groups that provide fitness advantages. Many polymer acylation pathways involve pairs of membrane-bound O-acyltransferase (MBOAT) and SGNH family proteins. As an example, the MBOAT protein PatA and the SGNH protein PatB are required in Gram-negative bacteria for peptidoglycan O-acetylation. The mechanism for how MBOAT-SGNH transferases move acyl groups from acyl-CoA donors made in the cytoplasm to extracellular polymers is unclear. Using the peptidoglycan O-acetyltransferase proteins PatAB, we explore the mechanism of MBOAT-SGNH pairs. We find that the MBOAT protein PatA catalyzes auto-acetylation of an invariant Tyr residue in its conserved C-terminal hexapeptide motif. We also show that PatB can use a synthetic hexapeptide containing an acetylated tyrosine to donate an acetyl group to a peptidoglycan mimetic. Finally, we report the structure of PatB, finding that it has structural features that shape its activity as an O-acetyltransferase and distinguish it from other SGNH esterases and hydrolases. Taken together, our results support a model for peptidoglycan acylation in which a tyrosine-containing peptide at the MBOAT's C-terminus shuttles an acyl group from the MBOAT active site to the SGNH active site, where it is transferred to peptidoglycan. This model likely applies to other systems containing MBOAT-SGNH pairs, such as those that O-acetylate alginate, cellulose, and secondary cell wall polysaccharides.
Keywords: O-acetyltransferase; X-ray crystallography; bacterial cell wall; o-acetylation; peptidoglycan.
Copyright © 2025 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare they have no conflicts of interest with the contents of this article.
Figures



Update of
-
The mechanism of peptidoglycan O-acetylation in Gram-negative bacteria typifies bacterial MBOAT-SGNH acyltransferases.bioRxiv [Preprint]. 2024 Sep 19:2024.09.17.613324. doi: 10.1101/2024.09.17.613324. bioRxiv. 2024. Update in: J Biol Chem. 2025 Jun;301(6):108531. doi: 10.1016/j.jbc.2025.108531. PMID: 39345430 Free PMC article. Updated. Preprint.
References
-
- Vollmer W., Blanot D., De Pedro M.A. Peptidoglycan structure and architecture. FEMS Microbiol. Rev. 2008;32:149–167. - PubMed
-
- Whitfield C., Trent M.S. Biosynthesis and export of bacterial lipopolysaccharides. Annu. Rev. Biochem. 2014;83:99–128. - PubMed
-
- Bera A., Herbert S., Jakob A., Vollmer W., Götz F. Why are pathogenic staphylococci so lysozyme resistant? The peptidoglycan O-acetyltransferase OatA is the major determinant for lysozyme resistance of Staphylococcus aureus. Mol. Microbiol. 2005;55:778–787. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

