Genetic variability, genotyping, and genomics of Mycobacterium leprae
- PMID: 40280733
- PMCID: PMC12035532
- DOI: 10.1093/femsre/fuaf012
Genetic variability, genotyping, and genomics of Mycobacterium leprae
Abstract
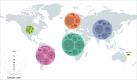
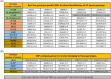
Leprosy, caused by Mycobacterium leprae and Mycobacterium lepromatosis, remains a significant global health issue despite a tremendous decline in its worldwide prevalence in the last four decades. Mycobacterium leprae strains possess very limited genetic variability, making it difficult to distinguish them using traditional genotyping tools. Successful genome sequencing of a considerable number of M. leprae strains in the recent past has allowed development of improved genotyping tools for the molecular epidemiology of leprosy. Comparative genomics has identified distinct M. leprae genotypes and revealed their characteristic genomic markers. This review summarizes the progress made in M. leprae genomics, with special emphasis on the development of genotyping schemes. Further, an updated genotyping scheme is introduced that also includes the newly reported genotypes 1B_Bangladesh, 1D_Malagasy, 3K-0/3K-1, 3Q and 4N/O. Additionally, genotype-specific markers (single nucleotide polymorphisms, Insertion/Deletion) have been incorporated into the typing scheme for the first time to enable differentiation of closely related strains. This will be particularly useful for geographic regions where M. leprae strains characterized by a small number of genotypes are predominant. The detailed compilation of genomic markers will also enable accurate identification of M. leprae genotypes, using targeted analysis of variable regions. Such markers are good candidates for developing artificial intelligence-based algorithms for classifying M. leprae genomic datasets.
Keywords: Mycobacterium leprae; comparative genomics; genetic diversity; genomic variability; genotyping; phylogeography.
© The Author(s) 2025. Published by Oxford University Press on behalf of FEMS.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures




References
-
- Avanzi C, Busso P, Benjak A et al. Transmission of drug-resistant leprosy in Guinea-Conakry detected using molecular epidemiological approaches. Clin Infect Dis. 2016;63:1482–4. - PubMed
-
- Avanzi C, del-Pozo J, Benjak A et al. Red squirrels in the British Isles are infected with leprosy bacilli. Science. 2016;354:744–7. - PubMed
-
- Avanzi C, Singh P, Truman RW et al. Molecular epidemiology of leprosy: an update. Infect Genet Evol. 2020;86:104581. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

