Unravelling the key factors for the dominance of Leuconostoc starters during kimchi fermentation
- PMID: 40280915
- PMCID: PMC12032216
- DOI: 10.1038/s41538-025-00415-w
Unravelling the key factors for the dominance of Leuconostoc starters during kimchi fermentation
Abstract
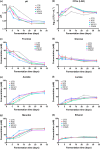
Recent studies aim to prevent kimchi spoilage and enhance the sensory and nutritional qualities using lactic acid bacteria, particularly Leuconostoc species, as kimchi starters. However, the factors enabling the successful adaptation and predominance of Leuconostoc species remain unclear. This study investigates the factors that contribute to the successful adaptation of Leuconostoc starter strains WiKim32, WiKim33, WiKim0121 and CBA3628 during kimchi fermentation using a comprehensive multi-omics approach. Our findings reveal that ATP-dependent molecular chaperones, which respond to cold and acidic kimchi environments, play crucial roles in successfully adapting Leuconostoc starter strains. Moreover, genes involved in carbohydrate metabolic pathways enhance ATP production, thereby supporting chaperone activity and bacterial growth. This study highlights the practical use of Leuconostoc starter strains WiKim32, WiKim33 and WiKim0121 and identifies essential factors for their successful adaptation and predominance during kimchi fermentation.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures






References
-
- Cha, J. et al. Does kimchi deserve the status of a probiotic food?. Crit. Rev. Food Sci. Nutr.64, 6512–6525 (2024). - PubMed
-
- Jung, M. Y. et al. Role of jeotgal, a Korean traditional fermented fish sauce, in microbial dynamics and metabolite profiles during kimchi fermentation. Food Chem.265, 135–143 (2018). - PubMed
-
- Kim, H. J. et al. A review of the health benefits of kimchi functional compounds and metabolites. Microbiol. Biotechnol. Lett.51, 353–373 (2023).
-
- Lee, S. H., Whon, T. W., Roh, S. W. & Jeon, C. O. Unraveling microbial fermentation features in kimchi: from classical to meta-omics approaches. Appl. Microbiol. Biotechnol.104, 7731–7744 (2020). - PubMed
-
- Lee, S. H., Jung, J. Y. & Jeon, C. O. Source tracking and succession of kimchi lactic acid bacteria during fermentation. J. Food Sci.80, M1871–M1877 (2015). - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

