IL-17A-producing NKp44(-) group 3 innate lymphoid cells accumulate in Familial Adenomatous Polyposis duodenal tissue
- PMID: 40280932
- PMCID: PMC12032359
- DOI: 10.1038/s41467-025-58907-y
IL-17A-producing NKp44(-) group 3 innate lymphoid cells accumulate in Familial Adenomatous Polyposis duodenal tissue
Abstract
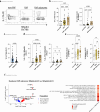
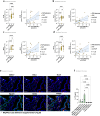
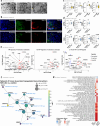
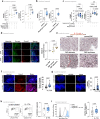
Familial adenomatous polyposis (FAP) is an inherited gastrointestinal syndrome associated with duodenal adenoma formation. Even among carriers of the same genetic variant, duodenal phenotypes vary, indicating that additional factors, such as the local immune system, play a role. We observe an increase in duodenal IL-17A(+)NKp44(-) innate lymphoid type 3 cell (ILC3) in FAP, localized near the epithelium and enriched in adenomas and carcinomas. Elevated IL1B, IL23A, and DLL4 transcript levels correlate with IL-17A(+)NKp44(-)ILC3 accumulation, and in vitro studies with duodenal organoids confirmed this relationship. Bulk RNA sequencing reveals upregulated Reactive oxygen species (ROS)-inducing enzymes DUOX2 and DUOXA2 in FAP adenomas. IL-17A-stimulated FAP organoids show increased DUOX2/DUOXA2 expression, Duox2 protein, and ROS production, leading to DNA damage, suggesting a mechanism by which these immune cells promote tumorigenesis. These findings suggest IL-17A(+)NKp44(-)ILC3s may contribute to a local environment that makes the epithelium more submissive for oncogenic transformation in FAP.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: R.H. and J.N. received endoscopic equipment on loan from Fujifilm, Germany. Robert Hüneburg has received consulting fees for medical advice from CPP-FAP, One Two Therapeutics, and Janssen Pharmaceuticals. C.P.S. has received speaker honoraria from Falk Pharma, Eisai, Astellas, Chiesi, MSD, and support for seminars from Gilead, Bristol-Myers Squibb, Abbvie, MSD, Norgine, Tillots Pharma, Eisai, Janssen, Falk Foundation, and consulting fees for medical advice from Fa. Schwabe, Astra Zeneca, Eisai and Astellas. The remaining authors declare no competing interests.
Figures




References
MeSH terms
Substances
Grants and funding
- SPP1937/Deutsche Forschungsgemeinschaft (German Research Foundation)
- 390873048/Deutsche Forschungsgemeinschaft (German Research Foundation)
- 432325352/Deutsche Forschungsgemeinschaft (German Research Foundation)
- KR 4521/1-1/Deutsche Forschungsgemeinschaft (German Research Foundation)
- SFB 1444 P14/Deutsche Forschungsgemeinschaft (German Research Foundation)
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

