Untargeted lipidomics profiling provides novel insights into pediatric patients with sepsis: an exploratory study
- PMID: 40281388
- PMCID: PMC12031908
- DOI: 10.1007/s11306-025-02255-x
Untargeted lipidomics profiling provides novel insights into pediatric patients with sepsis: an exploratory study
Abstract
Introduction: The plasma lipidome has emerged as an important indicator for assessing host metabolic and immune status in sepsis. While previous studies have largely examined specific lipid class changes in adults sepsis, comprehensive investigations into plasma lipidomic alterations in pediatric sepsis are limited. This study aimed to characterize the plasma lipidome in pediatric sepsis using a metabolomics-based exploratory approach, providing insights into pathophysiological mechanisms and potential biomarkers.
Methods: A retrospective study was conducted on pediatric patients with sepsis admitted to the pediatric intensive care unit (PICU). Untargeted lipidomics analysis using ultra-performance liquid chromatography coupled with Orbitrap mass spectrometry (UPLC-Orbitrap) was performed to compare metabolomic profiles between non-infected control patients and sepsis patients.
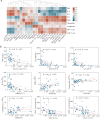
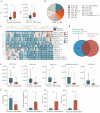
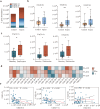
Results: Compared to controls, plasma lipid levels in sepsis patients decreased by 33.3%, increased by 20.2%, and remained unchanged in 46.5% of cases. A total of 1,257 differential lipids were identified in sepsis patients, with 24 lipids showing significant associations with pSOFA scores. In the recovery and deterioration subgroups, 186 differential lipids were identified, with triglyceride (TG) representing the highest proportion at 16.4%. Notably, 15 lipids with significant statistical differences were identified as differential lipid species through a comparison of those associated with pSOFA scores and those linked to sepsis prognosis. Fatty acid (FA) levels were significantly elevated in the sepsis group compared to controls, with arachidonic acid (FA(20:4)) showing the most significant increase (P < 0.001).
Conclusion: Alterations in plasma lipid profiles among children with sepsis reflect disease severity, systemic inflammatory responses, and sepsis prognosis. These findings underscore the prognostic potential of lipidomics and its value in understanding sepsis pathophysiology.
Keywords: Fatty acid; Pediatric sepsis; Plasma lipidome; Untargeted lipidomics.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests. Ethical approval and consent to participate: Ethical approval for the study was provided by Ethics Committee of Children’s Hospital of Fudan University ([2024] No. 116). Written informed consent was obtained from all parents or their surrogates of studied children. Consent for publication: All authors consent to the publication of this manuscript.
Figures


Similar articles
-
Systemic Inflammatory Response Syndrome.2025 Jun 20. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2025 Jun 20. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 31613449 Free Books & Documents.
-
Serum Ferritin Levels as a Prognostic Marker for Predicting Outcomes in Children With Severe Sepsis and Their Correlation With Pediatric Sequential Organ Failure Assessment Score.Cureus. 2025 May 19;17(5):e84436. doi: 10.7759/cureus.84436. eCollection 2025 May. Cureus. 2025. PMID: 40539133 Free PMC article.
-
Plasma metabolites associated with endometriosis in adolescents and young adults.Hum Reprod. 2025 May 1;40(5):843-854. doi: 10.1093/humrep/deaf040. Hum Reprod. 2025. PMID: 40107296
-
Psychological and/or educational interventions for the prevention of depression in children and adolescents.Cochrane Database Syst Rev. 2004;(1):CD003380. doi: 10.1002/14651858.CD003380.pub2. Cochrane Database Syst Rev. 2004. Update in: Cochrane Database Syst Rev. 2011 Dec 07;(12):CD003380. doi: 10.1002/14651858.CD003380.pub3. PMID: 14974014 Updated.
-
The Black Book of Psychotropic Dosing and Monitoring.Psychopharmacol Bull. 2024 Jul 8;54(3):8-59. Psychopharmacol Bull. 2024. PMID: 38993656 Free PMC article. Review.
References
-
- Balamuth, F., Scott, H. F., Weiss, S. L., Webb, M., Chamberlain, J. M., Bajaj, L., Depinet, H., Grundmeier, R. W., Campos, D., Deakyne Davies, S. J., Simon, N. J., Cook, L. J., & Alpern, E. R. (2022). Validation of the pediatric sequential organ failure assessment score and evaluation of third international consensus definitions for Sepsis and septic shock definitions in the pediatric emergency department. JAMA Pediatr, 176(7), 672–678. 10.1001/jamapediatrics.2022.1301 - PMC - PubMed
-
- Barber, G., Tanic, J., & Leligdowicz, A. (2023). Circulating protein and lipid markers of early sepsis diagnosis and prognosis: A scoping review. Current Opinion in Lipidology, 34(2), 70–81. 10.1097/mol.0000000000000870 - PubMed
-
- Bauer, M., Gerlach, H., Vogelmann, T., Preissing, F., Stiefel, J., & Adam, D. (2020). Mortality in sepsis and septic shock in Europe, North America and Australia between 2009 and 2019- results from a systematic review and meta-analysis. Critical Care, 24(1), 239. 10.1186/s13054-020-02950-2 - PMC - PubMed
-
- Cambiaghi, A., Díaz, R., Martinez, J. B., Odena, A., Brunelli, L., Caironi, P., Masson, S., Baselli, G., Ristagno, G., Gattinoni, L., de Oliveira, E., Pastorelli, R., & Ferrario, M. (2018). An innovative approach for the integration of proteomics and metabolomics data in severe septic shock patients stratified for mortality. Scientific Reports, 8(1), 6681. 10.1038/s41598-018-25035-1 - PMC - PubMed
-
- Caterino, M., Gelzo, M., Sol, S., Fedele, R., Annunziata, A., Calabrese, C., Fiorentino, G., D’Abbraccio, M., Dell’Isola, C., Fusco, F. M., Parrella, R., Fabbrocini, G., Gentile, I., Andolfo, I., Capasso, M., Costanzo, M., Daniele, A., Marchese, E., Polito, R., Russo, R., Missero, C., Ruoppolo, M., & Castaldo, G. (2021). Dysregulation of lipid metabolism and pathological inflammation in patients with COVID-19. Scientific Reports, 11(1), 2941. 10.1038/s41598-021-82426-7 - PMC - PubMed
MeSH terms
Substances
Grants and funding
- 2023ZDFC0103/Municipal Health System Key Supporting Discipline Project
- PPXK2024-06/Ningbo Medical and Health Brand Discipline
- 2021YFC2701800/National Key Research and Development Program of China
- ZD2021CY001/Shanghai Municipal Science and Technology Major Project
- 2308085MH267/Natural Science Foundation of Anhui Province
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
