Comprehensive analysis of the NAC transcription factor gene family in Sophora tonkinensis Gagnep
- PMID: 40281421
- PMCID: PMC12023634
- DOI: 10.1186/s12870-025-06564-0
Comprehensive analysis of the NAC transcription factor gene family in Sophora tonkinensis Gagnep
Abstract
Background: Sophora tonkinensis Gagnep. has long been utilized in the treatment of anti-inflammatory and pain-relieving, with its principal active compounds being alkaloids and flavonoids. NAC transcription factors, a large family of plant-specific regulators, play pivotal roles in growth, development, stress responses, and secondary metabolism. However, comprehensive genome-wide characterization of S. tonkinensis NAC gene family (StNAC) remains unexplored.
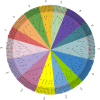
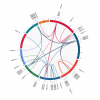
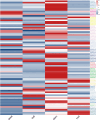
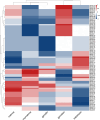
Results: This study identified 85 NAC proteins from the S. tonkinensis genome database. Phylogenetic analysis revealed that StNAC proteins were categorized into 15 subgroups based on their homology with Arabidopsis thaliana NAC proteins. Gene structure analysis demonstrated a variation in intron numbers ranging from 1 to 7, with a majority of StNAC genes containing 2-3 introns. Chromosomal distribution analysis indicated an uneven spread of StNAC genes across 9 chromosomes, with the highest number of StNAC genes on Chr3. Detection of 4 tandem duplicates and 32 segmental duplicates revealed that segmental duplication primarily drive StNAC genes amplification. Prediction of cis-regulatory elements suggested the involvement of StNAC genes in growth, stress responses, and hormone regulation. Gene expression analysis showed substantial variability expression of StNAC genes across different tissues. Notably, eight StNAC genes were identified as significantly associated alkaloid and flavonoid levels. qRT-PCR validation indicated that five genes were highly expressed in tissues, corroborating transcriptome data.
Conclusion: These findings offer valuable insights for further functional characterization of NAC genes and their potential roles in alkaloid and flavonoid biosynthesis in S. tonkinensis.
Keywords: Sophora tonkinensis Gagnep; Alkaloid; Flavonoid; Gene family; NAC transcription factor.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures




Similar articles
-
Genes involved in the regulation of alkaloid and flavonoid biosynthesis in different tissues of Sophora tonkinensis via transcriptomics and metabolomics.BMC Plant Biol. 2025 Jul 2;25(1):840. doi: 10.1186/s12870-025-06865-4. BMC Plant Biol. 2025. PMID: 40604440 Free PMC article.
-
Genome-wide organization and expression profiling of the NAC transcription factor family in potato (Solanum tuberosum L.).DNA Res. 2013 Aug;20(4):403-23. doi: 10.1093/dnares/dst019. Epub 2013 May 5. DNA Res. 2013. PMID: 23649897 Free PMC article.
-
Integrated transcriptome and small RNA sequencing analyses reveal a drought stress response network in Sophora tonkinensis.BMC Plant Biol. 2021 Dec 2;21(1):566. doi: 10.1186/s12870-021-03334-6. BMC Plant Biol. 2021. PMID: 34856930 Free PMC article.
-
Genome-Wide Identification and Analysis of the NAC Transcription Factor Gene Family in Garden Asparagus (Asparagus officinalis).Genes (Basel). 2022 May 30;13(6):976. doi: 10.3390/genes13060976. Genes (Basel). 2022. PMID: 35741738 Free PMC article.
-
Genome-wide identification, tissue expression pattern, and salt stress response analysis of the NAC gene family in Thinopyrum elongatum.BMC Plant Biol. 2025 May 15;25(1):643. doi: 10.1186/s12870-025-06696-3. BMC Plant Biol. 2025. PMID: 40375071 Free PMC article.
References
-
- Souer E, van Houwelingen A, Kloos D, Mol J, Koes R. The no apical meristem gene of Petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries. Cell. 1996;85(2):159–70. - PubMed
-
- Kikuchi K, Ueguchi-Tanaka M, Yoshida KT, Matsusoka M, Hirano HY. Molecular analysis of the NAC gene family in rice. Mol Gen Genet. 2000;262(6):1047–51. - PubMed
-
- Olsen AN, Ernst HA, Lo Leggio L, Skriver K. NAC transcription factors: structurally distinct, functionally diverse. Trends Plant Sci. 2005;10(2):79–87. - PubMed
-
- Luo G, Yang Y, Zhou M, Ye Q, Liu Y, Gu J, et al. Novel 2-arylbenzofuran dimers and polyisoprenylated flavanones from Sophora tonkinensis. Fitoterapia. 2014;99:21–7. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

