Plasma cell-free DNA methylation profile before afatinib treatment is associated with progression-free and overall survival of patients with epidermal growth factor receptor gene mutation-positive non-small cell lung cancer
- PMID: 40281631
- PMCID: PMC12032777
- DOI: 10.1186/s13148-025-01870-8
Plasma cell-free DNA methylation profile before afatinib treatment is associated with progression-free and overall survival of patients with epidermal growth factor receptor gene mutation-positive non-small cell lung cancer
Abstract
Background: The present study aimed to clarify the clinical significance of the cell-free DNA (cfDNA) methylation profile of patients with non-small cell lung cancer (NSCLC) showing the epidermal growth factor receptor (EGFR) gene mutation.
Methods: In 103 patients, genome-wide DNA methylation analysis using Infinium Methylation EPIC array was performed using samples of pre-tyrosine kinase inhibitor afatinib-treatment plasma cfDNA (n = 101) and post-afatinib cfDNA (n = 84).
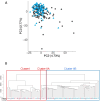
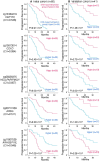
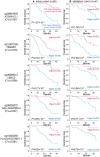
Results: Principal component analysis indicated that the cfDNA methylation profile was altered after afatinib treatment. Hierarchical clustering using the pre-afatinib cfDNA methylation profile revealed that cases with a fatal outcome were accumulated in specific clusters. Moreover, Kaplan-Meier analysis showed that the pre-afatinib cfDNA methylation profile was significantly associated with both progression-free survival (PFS) and overall survival (OS), whereas the post-afatinib profile was not. The genes for which pre-afatinib cfDNA methylation levels were associated with PFS were accumulated in the cadherin, Wnt, and EGFR signaling pathways. Activation of EGFR-related signaling due to DNA methylation alterations might overturn the effect of afatinib. Pre-afatinib levels of CEP170 and CHCHD6 cfDNA methylation were associated with both PFS and OS. Both pre- and post-afatinib cfDNA methylation levels of SLC9A3R2 and INTS1 were associated with bone metastasis. Using the cfDNA methylation levels at two CpG sites, cg12721600 and cg05905155, patients showing an overall response were predicted with a sensitivity of 96% or more.
Conclusions: The non-invasively measurable cfDNA methylation profile may reflect the corresponding profile in cancer cells, and that pre-treatment measurement may provide clinically useful information on EGFR mutation-positive NSCLC.
Keywords: Afatinib; Cell-free DNA; DNA methylation; Epidermal growth factor receptor; Lung adenocarcinoma.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: This study complied with the Declaration of Helsinki and was approved by the Ethics Committee at Keio University School of Medicine (2021–0147) and the Ethical Review Board for Medical and Health Research Involving Human Subjects at Teikyo University (16–066). All participants provided written informed consent. Competing interests: AT is an employee of Nippon Boehringer Ingelheim Co., Ltd., Tokyo, Japan.
Figures
Similar articles
-
First-line treatment of advanced epidermal growth factor receptor (EGFR) mutation positive non-squamous non-small cell lung cancer.Cochrane Database Syst Rev. 2021 Mar 18;3(3):CD010383. doi: 10.1002/14651858.CD010383.pub3. Cochrane Database Syst Rev. 2021. PMID: 33734432 Free PMC article.
-
Best Response According to RECIST During First-line EGFR-TKI Treatment Predicts Survival in EGFR Mutation-positive Non-Small-cell Lung Cancer Patients.Clin Lung Cancer. 2018 May;19(3):e361-e372. doi: 10.1016/j.cllc.2018.01.005. Epub 2018 Feb 1. Clin Lung Cancer. 2018. PMID: 29477365
-
Feasibility and effectiveness of afatinib for poor performance status patients with EGFR-mutation-positive non-small-cell lung cancer: a retrospective cohort study.BMC Cancer. 2021 Jul 27;21(1):859. doi: 10.1186/s12885-021-08587-w. BMC Cancer. 2021. PMID: 34315431 Free PMC article.
-
[Influence of Different Therapies on EGFR Mutants by Circulating Cell-free DNA of Lung Adenocarcinoma and Prognosis].Zhongguo Fei Ai Za Zhi. 2018 May 20;21(5):389-396. doi: 10.3779/j.issn.1009-3419.2018.05.06. Zhongguo Fei Ai Za Zhi. 2018. PMID: 29764589 Free PMC article. Chinese.
-
Afatinib as First-Line Treatment in Asian Patients with EGFR Mutation-Positive NSCLC: A Narrative Review of Real-World Evidence.Adv Ther. 2021 May;38(5):2038-2053. doi: 10.1007/s12325-021-01696-9. Epub 2021 Mar 17. Adv Ther. 2021. PMID: 33730350 Free PMC article. Review.
References
-
- Jones PA, Issa J-P, Baylin S. Targeting the cancer epigenome for therapy. Nat Rev Genet. 2016;17:630–41. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

