Pathogenomic Insights into Piscirickettsia salmonis with a Focus on Virulence Factors, Single-Nucleotide Polymorphism Identification, and Resistance Dynamics
- PMID: 40282010
- PMCID: PMC12024244
- DOI: 10.3390/ani15081176
Pathogenomic Insights into Piscirickettsia salmonis with a Focus on Virulence Factors, Single-Nucleotide Polymorphism Identification, and Resistance Dynamics
Abstract
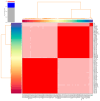
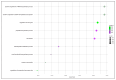
Effective control of bacterial infections remains a significant challenge in aquaculture. The marine bacterium Piscirickettsia salmonis (P. salmonis), responsible for piscirickettsiosis, causes widespread infections in various salmon species, leading to substantial mortality and economic losses. Despite efforts to genetically characterize P. salmonis, critical gaps persist in understanding its virulence factors, antimicrobial resistance genes, and single-nucleotide polymorphisms (SNPs). This study addresses these gaps through a comparative analysis of the pan-genome and core genomes of 80 P. salmonis strains from different geographical regions and genogroups. P. salmonis had an open pan-genome consisting of 14,564 genes, with a core genome of 1257 conserved genes. Eleven virulence-related genes were identified in the pan-genome, categorized into five functional groups, providing new insights into the pathogenicity of P. salmonis. Unique SNPs were detected in four key genes (gyrA, dnaK, rpoB, and ftsZ), serving as robust molecular markers for distinguishing the LF and EM genogroups. Notably, AMR genes identified in four LF strains suggest evolutionary adaptations under selective pressure. Functional annotation of the core genomes using the gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases demonstrated conserved gene clusters linked to essential intracellular survival mechanisms and bacterial pathogenicity. These findings suggest a direct association between core genome features and variations in pathogenesis and host-pathogen interactions across genogroups. Phylogenetic reconstruction further highlighted the influence of AMR genes on strain divergence. Collectively, this study enhances the genomic understanding of P. salmonis and lays the groundwork for improved diagnostic tools and targeted therapeutics to manage piscirickettsiosis in aquaculture.
Keywords: SNPs; genomics; pathogenesis; piscirickettsiosis; virulence.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




References
-
- Food and Agriculture Organization of the United Nations (FAO) The State of World Fisheries and Aquaculture 2022. FAO; Rome, Italy: 2022. - DOI
-
- Marinho-Neto F.A., Claudiano G.S., Yunis-Aguinaga J., Cueva-Quiroz V.A., Kobashigawa K.K., Cruz N.R., Moraes F.R., Moraes J.R. Morphological, microbiological and ultrastructural aspects of sepsis by Aeromonas hydrophila in Piaractus mesopotamicus. PLoS ONE. 2019;14:e0222626. doi: 10.1371/journal.pone.0222626. - DOI - PMC - PubMed
-
- Iversen A., Asche F., Hermansen Ø., Nystøyl R. Production cost and competitiveness in major salmon farming countries 2003–2018. Aquaculture. 2020;522:735089. doi: 10.1016/j.aquaculture.2020.735089. - DOI
LinkOut - more resources
Full Text Sources

