Biomolecules Interacting with Long Noncoding RNAs
- PMID: 40282307
- PMCID: PMC12025117
- DOI: 10.3390/biology14040442
Biomolecules Interacting with Long Noncoding RNAs
Abstract
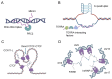
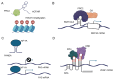
This review explores the complex interactions between long noncoding RNAs (lncRNAs) and other biomolecules, highlighting their pivotal roles in gene regulation and cellular function. LncRNAs, defined as RNA transcripts exceeding 200 nucleotides without encoding proteins, are involved in diverse biological processes, from embryogenesis to pathogenesis. They interact with DNA through mechanisms like triplex structure formation, influencing chromatin organization and gene expression. LncRNAs also modulate RNA-mediated processes, including mRNA stability, translational control, and splicing regulation. Their versatility stems from their forming of complex structures that enable interactions with various biomolecules. This review synthesizes current knowledge on lncRNA functions, discusses emerging roles in development and disease, and evaluates potential applications in diagnostics and therapeutics. By examining lncRNA interactions, it provides insights into the intricate regulatory networks governing cellular processes, underscoring the importance of lncRNAs in molecular biology. Unlike the majority of previous reviews that primarily focused on individual aspects of lncRNA biology, this comprehensive review uniquely integrates structural, functional, and mechanistic perspectives on lncRNA interactions across diverse biomolecules. Additionally, this review critically evaluates cutting-edge methodologies for studying lncRNA interactions, bridges fundamental molecular mechanisms with potential clinical applications, and highlights their potential.
Keywords: RNA–DNA triplexes; cellular function; gene regulation; long noncoding RNA; miRNA sponge.
Conflict of interest statement
The author declares no conflicts of interest.
Figures

Similar articles
-
Genome-wide computational analysis of potential long noncoding RNA mediated DNA:DNA:RNA triplexes in the human genome.J Transl Med. 2017 Sep 2;15(1):186. doi: 10.1186/s12967-017-1282-9. J Transl Med. 2017. PMID: 28865451 Free PMC article.
-
LncmiRSRN: identification and analysis of long non-coding RNA related miRNA sponge regulatory network in human cancer.Bioinformatics. 2018 Dec 15;34(24):4232-4240. doi: 10.1093/bioinformatics/bty525. Bioinformatics. 2018. PMID: 29955818
-
Cytoplasmic functions of long noncoding RNAs.Wiley Interdiscip Rev RNA. 2018 May;9(3):e1471. doi: 10.1002/wrna.1471. Epub 2018 Mar 8. Wiley Interdiscip Rev RNA. 2018. PMID: 29516680 Free PMC article. Review.
-
RNA-DNA Triplex Formation by Long Noncoding RNAs.Cell Chem Biol. 2016 Nov 17;23(11):1325-1333. doi: 10.1016/j.chembiol.2016.09.011. Epub 2016 Oct 20. Cell Chem Biol. 2016. PMID: 27773629 Review.
-
Comprehensive analysis of long noncoding RNA (lncRNA)-chromatin interactions reveals lncRNA functions dependent on binding diverse regulatory elements.J Biol Chem. 2019 Oct 25;294(43):15613-15622. doi: 10.1074/jbc.RA119.008732. Epub 2019 Sep 4. J Biol Chem. 2019. PMID: 31484726 Free PMC article.
Cited by
-
LncRNAs regulates cell death in osteosarcoma.Sci Rep. 2025 Jul 2;15(1):22592. doi: 10.1038/s41598-025-04440-3. Sci Rep. 2025. PMID: 40595878 Free PMC article.
References
-
- Mattick J.S., Amaral P.P., Carninci P., Carpenter S., Chang H.Y., Chen L.L., Chen R., Dean C., Dinger M.E., Fitzgerald K.A., et al. Long non-coding RNAs: Definitions, functions, challenges and recommendations. Nat. Rev. Mol. Cell Biol. 2023;24:430–447. doi: 10.1038/s41580-022-00566-8. - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources

