Whole-Genome Resequencing in Sheep: Applications in Breeding, Evolution, and Conservation
- PMID: 40282323
- PMCID: PMC12026845
- DOI: 10.3390/genes16040363
Whole-Genome Resequencing in Sheep: Applications in Breeding, Evolution, and Conservation
Abstract
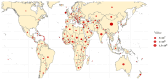
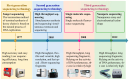
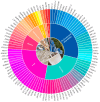
Sheep (Ovis aries) were domesticated around 10,000 years ago and have since become an integral part of human agriculture, providing essential resources, such as wool, meat, and milk. Over the past century, advances in communication and agricultural productivity have driven the evolution of selective breeding practices, further enhancing the value of sheep in the global economy. Recently, the rapid development of whole-genome resequencing (WGR) technologies has significantly accelerated research in sheep molecular biology, facilitating the discovery of genetic underpinnings for critical traits. This review offers a comprehensive overview of the evolution of whole-genome resequencing and its application to sheep genetics. It explores the domestication and genetic origins of sheep, examines the genetic structure and differentiation of various sheep populations, and discusses the use of WGR in the development of genetic maps. In particular, the review highlights how WGR technology has advanced our understanding of key traits, such as wool production, lactation, reproductive performance, disease resistance, and environmental adaptability. The review also covers the use of WGR technology in the conservation and sustainable utilization of sheep genetic resources, offering valuable insights for future breeding programs aimed at enhancing the genetic diversity and resilience of sheep populations.
Keywords: genetic map; genetic resources; population genetic differentiation; sheep; trait association analysis; whole-genome resequencing.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
Similar articles
-
Construction of a high-density genetic map and QTL localization of body weight and wool production related traits in Alpine Merino sheep based on WGR.BMC Genomics. 2024 Jun 27;25(1):641. doi: 10.1186/s12864-024-10535-4. BMC Genomics. 2024. PMID: 38937677 Free PMC article.
-
Population structure and selection signal analysis of indigenous sheep from the southern edge of the Taklamakan Desert.BMC Genomics. 2024 Jul 9;25(1):681. doi: 10.1186/s12864-024-10581-y. BMC Genomics. 2024. PMID: 38982349 Free PMC article.
-
Genetic diversity and population structure of Tibetan sheep breeds determined by whole genome resequencing.Trop Anim Health Prod. 2021 Feb 20;53(1):174. doi: 10.1007/s11250-021-02605-6. Trop Anim Health Prod. 2021. PMID: 33611716
-
The evolution of contemporary livestock species: Insights from mitochondrial genome.Gene. 2024 Nov 15;927:148728. doi: 10.1016/j.gene.2024.148728. Epub 2024 Jun 27. Gene. 2024. PMID: 38944163 Review.
-
Genetics of the phenotypic evolution in sheep: a molecular look at diversity-driving genes.Genet Sel Evol. 2022 Sep 9;54(1):61. doi: 10.1186/s12711-022-00753-3. Genet Sel Evol. 2022. PMID: 36085023 Free PMC article. Review.
Cited by
-
Decoding the Function of FGFBP1 in Sheep Adipocyte Proliferation and Differentiation.Animals (Basel). 2025 May 18;15(10):1456. doi: 10.3390/ani15101456. Animals (Basel). 2025. PMID: 40427333 Free PMC article.
References
-
- Larsson M.N.A., Morell Miranda P., Pan L., Başak Vural K., Kaptan D., Rodrigues Soares A.E., Kivikero H., Kantanen J., Somel M., Özer F., et al. Ancient Sheep Genomes Reveal Four Millennia of North European Short-Tailed Sheep in the Baltic Sea Region. Genome Biol. Evol. 2024;16:evae114. doi: 10.1093/gbe/evae114. - DOI - PMC - PubMed
-
- Saadatabadi L.M., Mohammadabadi M., Ghanatsaman Z.A., Babenko O., Stavetska R.V., Kalashnik O.M., Afanasenko V., Kochuk-Yashchenko O.A., Kucher D.M., Nanaei H.A. Data of Whole-Genome Sequencing of Karakul, Zel, and Kermani Sheep Breeds. BMC Res. Notes. 2023;16:353. doi: 10.1186/s13104-023-06630-6. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

