Comparative Analysis of Salt Tolerance and Transcriptomics in Two Varieties of Agropyron desertorum at Different Developmental Stages
- PMID: 40282327
- PMCID: PMC12026692
- DOI: 10.3390/genes16040367
Comparative Analysis of Salt Tolerance and Transcriptomics in Two Varieties of Agropyron desertorum at Different Developmental Stages
Abstract
Background: Most of the grasslands in China are experiencing varying degrees of degradation, desertification, and salinization (collectively referred to as the "three degradations"), posing a serious threat to the country's ecological security. Agropyron desertorum, known for its wide distribution, strong adaptability, and resistance, is an excellent grass species for the ecological restoration of grasslands affected by the "three degradations". This study focused on two currently popular varieties of A. desertorum, exploring their salt tolerance mechanisms and identifying candidate genes for salt and alkali tolerance.
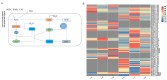
Methods: Transcriptome sequencing was performed on two varieties of A. desertorum during the seed germination and seedling stages under varying degrees of saline-alkali stress. At the seed stage, we measured the germination rate, relative germination rate, germination index, and salt injury rate under different NaCl concentrations. During the seedling stage, physiological indicators, including superoxide dismutase (SOD), peroxidase (POD), malondialdehyde (MDA), proline (PRO), soluble protein (SP), and catalase (CAT), were analyzed after exposure to 30, 60, 120, and 180 mM NaCl for 12 days. Analysis of differentially expressed genes (DEGs) at 6 and 24 h post-treatment with 120 mM NaCl revealed significant differences in the salt stress responses between the two cultivars.
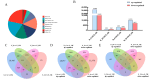
Results: Our study indicates that during the seed stage, A. desertorum (Schult.) exhibits a higher relative germination potential, relative germination rate, and relative germination index, along with a lower relative salt injury rate compared to A. desertorum cv. Nordan. Compared with A. desertorum cv. Nordan, A. desertorum (Schult.) has higher salt tolerance, which is related to its stronger antioxidant activity and different antioxidant-related pathways. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses were used to identify the key biological processes and pathways involved in salt tolerance, including plant hormone signal transduction, antioxidant defense, and cell membrane stability.
Conclusions: A. desertorum (Schult.) exhibits stronger salt tolerance than A. desertorum cv. Nordan. Salt stress at a concentration of 30-60 mM promotes the germination of the seeds of both Agropyron cultivars. The two Agropyron plants mainly overcome the damage caused by salt stress through the AsA-GSH pathway. This study provides valuable insights into the molecular mechanisms of salt tolerance in Agropyron species and lays the groundwork for future breeding programs aimed at improving salt tolerance in desert grasses.
Keywords: Agropyron desertorum; RNA-seq; physiological analysis; salt stress; seed germination; transcriptomics.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures





References
-
- Zhou T., Lv Q., Zhang L., Fan J., Wang T., Meng Y., Xia H., Ren X., Hu S. Converted paddy to upland in saline-sodic land could improve soil ecosystem multifunctionality by enhancing soil quality and alleviating microbial metabolism limitation. Sci. Total Environ. 2024;924:171707. doi: 10.1016/j.scitotenv.2024.171707. - DOI - PubMed
-
- Wang G., Gang N.I., Feng G., Burrill H.M., Jianfang L.I., Zhang J., Zhang F. Saline-alkali soil reclamation and utilization in China: Progress and prospects. Front. Agric. Sci. 2024;11:216–228.
-
- Johnson R.C. Salinity Resistance, Water Relations, and Salt Content of Crested and Tall Wheatgrass Accessions. Crop Sci. 1991;31:730–734.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

