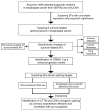
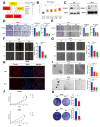
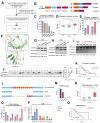
Multi-Omics Analysis of Survival-Related Splicing Factors and Identifies CRNKL1 as a Therapeutic Target in Esophageal Cancer
- PMID: 40282339
- PMCID: PMC12027253
- DOI: 10.3390/genes16040379
Multi-Omics Analysis of Survival-Related Splicing Factors and Identifies CRNKL1 as a Therapeutic Target in Esophageal Cancer
Abstract
Background: RNA alternative splicing represents a pivotal regulatory mechanism of eukaryotic gene expression, wherein splicing factors (SFs) serve as key regulators. Aberrant SF expression drives oncogenic splice variant production, thereby promoting tumorigenesis and malignant progression. However, the biological functions and potential targets of SFs remain largely underexplored. Methods: Through multi-omics analysis, we identified survival-related splicing factors (SFs) in esophageal cancer and elucidated their biological regulatory networks. To further investigate their downstream splicing targets, we combined alternative splicing events resulting from SF knockdown with those specific to esophageal cancer. Finally, these splicing events were validated through full-length RNA sequencing and confirmed in cancer cells and clinical specimens. Result: We identified six SFs that are highly expressed in esophageal cancer and correlate with poor prognosis. Further analysis revealed that these factors are significantly associated with immune infiltration, cancer stemness, tumor heterogeneity, and drug resistance. CRNKL1 was identified as a hub SFs. The target genes and pathways regulated by these SFs showed substantial overlap, suggesting their coordinated roles in promoting cancer stemness and metastasis. Specifically, alternative splicing of key markers, such as CD44 and CTTN, was regulated by most of these SFs and correlated with poor prognosis. Conclusions: Our study unveils six survival-related SFs that contribute to the aggressiveness of esophageal cancer and CTTN and CD44 alternative splicing may act as common downstream effectors of survival-related SFs. This study provides mechanistic insights into SF-mediated tumorigenesis and highlight novel therapeutic vulnerabilities in esophageal cancer.
Keywords: alternative splicing; esophageal cancer; splicing factors.
Conflict of interest statement
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures




Similar articles
-
Integrated multi-omics analysis of the clinical relevance and potential regulatory mechanisms of splicing factors in hepatocellular carcinoma.Bioengineered. 2021 Dec;12(1):3978-3992. doi: 10.1080/21655979.2021.1948949. Bioengineered. 2021. PMID: 34288818 Free PMC article.
-
Integrative analysis reveals the prognostic value and functions of splicing factors implicated in hepatocellular carcinoma.Sci Rep. 2021 Jul 26;11(1):15175. doi: 10.1038/s41598-021-94701-8. Sci Rep. 2021. PMID: 34312475 Free PMC article.
-
Study of prognostic splicing factors in cancer using machine learning approaches.Hum Mol Genet. 2024 Jun 21;33(13):1131-1141. doi: 10.1093/hmg/ddae047. Hum Mol Genet. 2024. PMID: 38538560 Free PMC article.
-
Splicing factors: Insights into their regulatory network in alternative splicing in cancer.Cancer Lett. 2021 Mar 31;501:83-104. doi: 10.1016/j.canlet.2020.11.043. Epub 2020 Dec 10. Cancer Lett. 2021. PMID: 33309781 Review.
-
The role of alternative splicing in cancer: From oncogenesis to drug resistance.Drug Resist Updat. 2020 Dec;53:100728. doi: 10.1016/j.drup.2020.100728. Epub 2020 Sep 28. Drug Resist Updat. 2020. PMID: 33070093 Review.
References
-
- Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68:394–424. - PubMed
-
- Ferlay J., Colombet M., Soerjomataram I., Mathers C., Parkin D.M., Pineros M., Znaor A., Bray F. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int. J. Cancer. 2019;144:1941–1953. - PubMed
-
- Gong L., Bai M., Dai L., Guo X., Leng X., Li Z., Lu Z., Mao T., Pang Q., Shen L., et al. CACA guidelines for holistic integrative management of esophageal carcinoma. Holist. Integr. Oncol. 2023;2:34.
-
- Xie S.-H., Huang R.-Q., Liu Y.-L., Cao S.-M., Qian C.-N. An increase in early cancer detection rates at a single cancer center: Experiences from Sun Yat-sen University Cancer Center. Vis. Cancer Med. 2022;3:1.
-
- Deboever N., Jones C.M., Yamashita K., Ajani J.A., Hofstetter W.L. Advances in diagnosis and management of cancer of the esophagus. BMJ. 2024;385:e074962. - PubMed
MeSH terms
Substances
Grants and funding
- No.81972785, No. 81773162 and No. 81572901 to B.H; No.31801037 to X.Z./National Natural Science Foundation of China
- No.2017A030313866, No. 2022A1515012298 and 2024A1515013084 to B.H/the Provincial Natural Science Foundation of Guangdong and Guangdong Basic and Applied Basic Research Foundation
- No. 410310543403, and No.2019JCZX05/the Science Foundation of Army Medical University
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

