Identification and Characterization of LINE and SINE Retrotransposons in the African Hedgehog (Atelerix albiventris, Erinaceidae) and Their Association with 3D Genome Organization and Gene Expression
- PMID: 40282356
- PMCID: PMC12026660
- DOI: 10.3390/genes16040397
Identification and Characterization of LINE and SINE Retrotransposons in the African Hedgehog (Atelerix albiventris, Erinaceidae) and Their Association with 3D Genome Organization and Gene Expression
Abstract
Background: The African hedgehog (Atelerix albiventris) exhibits specialized skin differentiation leading to spine formation, yet its regulatory mechanisms remain unclear. Transposable elements (TEs), particularly LINEs (long interspersed nuclear elements) and SINEs (short interspersed nuclear elements), are known to influence genome organization and gene regulation.
Objectives: Given the high proportion of SINEs in the hedgehog genome, this study aims to characterize the distribution, evolutionary dynamics, and potential regulatory roles of LINEs and SINEs, focusing on their associations with chromatin architecture, DNA methylation, and gene expression.
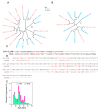
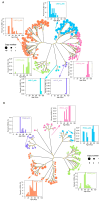
Methods: We analyzed LINE and SINE distribution using HiFi sequencing and classified TE families through phylogenetic reconstruction. Hi-C data were used to explore TE interactions with chromatin architecture, while whole-genome 5mCpG methylation was inferred from PacBio HiFi reads of muscle tissue using a deep-learning-based approach. RNA-seq data from skin tissues were analyzed to assess TE expression and potential associations with genes linked to spine development.
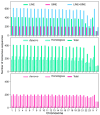
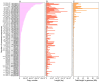
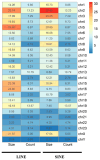
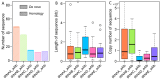
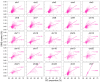
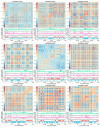
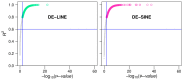
Results: SINEs form distinct genomic blocks in GC-rich and highly methylated regions, whereas LINEs are enriched in AT-rich, hypomethylated regions. LINEs and SINEs are associated differently with A/B compartments, with SINEs in euchromatin and LINEs in heterochromatin. Methylation analysis suggests that younger TEs tend to have higher methylation levels, and expression analysis indicates that some differentially expressed TEs may be linked to genes involved in epidermal and skeletal development.
Conclusions: This study provides a genome-wide perspective on LINE and SINE distribution, methylation patterns, and potential regulatory roles in A. albiventris. While not establishing a direct causal link, the findings suggest that TEs may influence gene expression associated with spine development, offering a basis for future functional studies.
Keywords: Atelerix albiventris; LINEs (long interspersed nuclear elements); SINEs (short interspersed nuclear elements); genome structure; repetitive sequences.
Conflict of interest statement
The authors declare no competing interests. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures


















References
-
- Santana E.M., Jantz H.E., Best T.L. Atelerix albiventris (Erinaceomorpha: Erinaceidae) Mamm. Species. 2010;42:99–110. doi: 10.1644/857.1. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

