GONNMDA: A Ordered Message Passing GNN Approach for miRNA-Disease Association Prediction
- PMID: 40282386
- PMCID: PMC12027447
- DOI: 10.3390/genes16040425
GONNMDA: A Ordered Message Passing GNN Approach for miRNA-Disease Association Prediction
Abstract
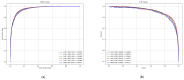
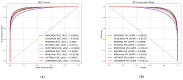
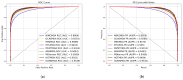
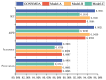
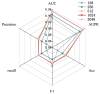
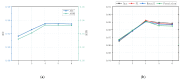
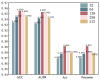
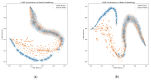
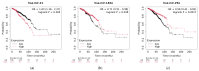
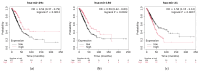
Small non-coding molecules known as microRNAs (miRNAs) play a critical role in disease diagnosis, treatment, and prognosis evaluation. Traditional wet-lab methods for validating miRNA-disease associations are often time-consuming and inefficient. With the advancement of high-throughput sequencing technologies, deep learning methods have become effective tools for uncovering potential patterns in miRNA-disease associations and revealing novel biological insights. Most of the existing approaches focus primarily on individual molecular behavior, overlooking interactions at the multi-molecular level. Conventional graph neural network (GNN) models struggle to generalize to heterogeneous graphs, and as network depth increases, node representations become indistinguishable due to over-smoothing, resulting in reduced predictive performance. GONNMDA first integrates similarity features from multiple data sources and applies noise reduction to obtain a reconstructed, comprehensive similarity representation. It then constructs heterogeneous graphs and applies a root-tree hierarchical alignment, along with an ordered gating message-passing mechanism, effectively addressing the challenges of heterogeneity and over-smoothing. Finally, a multilayer perceptron is employed to produce the final association predictions. To evaluate the effectiveness of GONNMDA, we conducted extensive experiments where the model achieved an AUC of 95.49% and an AUPR of 95.32%. The results demonstrate that GONNMDA outperforms several recent state-of-the-art methods. In addition, case studies and survival analyses on three common human cancers-breast cancer, rectal cancer, and lung cancer-further validate the effectiveness and reliability of GONNMDA in predicting miRNA-disease associations.
Keywords: heterogeneous graph; miRNA–disease association; ordered GNN; singular value decomposition.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




References
-
- Raponi M., Dossey L., Jatkoe T., Wu X., Chen G., Fan H., Beer D.G. MicroRNA classifiers for predicting prognosis of squamous cell lung cancer. Cancer Res. 2009;69:5776–5783. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

