Advancing Non-Invasive Prenatal Screening: A Targeted 1069-Gene Panel for Comprehensive Detection of Monogenic Disorders and Copy Number Variations
- PMID: 40282387
- PMCID: PMC12026569
- DOI: 10.3390/genes16040427
Advancing Non-Invasive Prenatal Screening: A Targeted 1069-Gene Panel for Comprehensive Detection of Monogenic Disorders and Copy Number Variations
Abstract
We introduce an innovative, non-invasive prenatal screening approach for detecting fetal monogenic alterations and copy number variations (CNVs) from maternal blood.
Method: Circulating free DNA (cfDNA) was extracted from maternal peripheral blood and processed using the VeriSeq NIPT Solution (Illumina, San Diego, CA, USA), with shallow whole-genome sequencing (sWGS) performed on a NextSeq550Dx (Illumina). A customized gene panel and bioinformatics tool, named the "VERA Revolution", were developed to detect variants and CNVs in cfDNA samples. Results were compared with genomic DNA (gDNA) extracted from fetal samples, including amniotic fluid and chorionic villus sampling and buccal swabs.
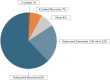
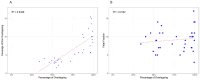
Results: The study included pregnant women with gestational ages from 10 + 3 to 15 + 2 weeks (mean: 12.1 weeks). The fetal fraction (FF), a crucial measure of cfDNA test reliability, ranged from 5% to 20%, ensuring adequate DNA amount for analysis. Among 36 families tested, 14 showed a wild-type genotype. Identified variants included two deletions (22q11.2, and 4p16.3), two duplications (16p13 and 5p15), and eighteen single-nucleotide variants (one in CFTR, three in GJB2, three in PAH, one in RIT1, one in DHCR7, one in TCOF1, one in ABCA4, one in MYBPC3, one in MCCC2, two in GBA1 and three in PTPN11). Significant concordance was found between our panel results and prenatal/postnatal genetic profiles.
Conclusions: The "VERA Revolution" test highlights advancements in prenatal genomic screening, offering potential improvements in prenatal care.
Keywords: circulating-free fetal DNA; copy number variations; monogenic disorders; next-generation sequencing; non-invasive prenatal screening.
Conflict of interest statement
Authors R.S., L.D.A., M.I., N.P., R.R., R.C., A.M., E.E., G.S., A.F. are employed by AMES. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Atkins J.C., Padgett C.R. Living with a Rare Disease: Psychosocial Impacts for Parents and Family Members—A Systematc Review. J. Child Fam. Stud. 2024;33:617–636. doi: 10.1007/s10826-024-02790-6. - DOI
-
- Zhang J., Li J., Saucier J.B., Feng Y., Jiang Y., Sinson J., McCombs A.K., Schmitt E.S., Peacock S., Chen S., et al. Non-Invasive Prenatal Sequencing for Multiple Mendelian Monogenic Disorders Using Circulating Cell-Free Fetal DNA. Nat. Med. 2019;25:439–447. doi: 10.1038/s41591-018-0334-x. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

