Identification and Expression Analysis of CCCH Zinc Finger Family Genes in Oryza sativa
- PMID: 40282389
- PMCID: PMC12026475
- DOI: 10.3390/genes16040429
Identification and Expression Analysis of CCCH Zinc Finger Family Genes in Oryza sativa
Abstract
Background: CCCH zinc finger proteins (OsC3Hs) are a class of transcriptional regulators that play important roles in plant development and stress responses. Although their functional significance has been widely studied in model species, comprehensive genome-wide characterization of CCCH proteins in rice (Oryza sativa) remains limited.
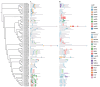
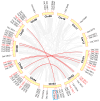
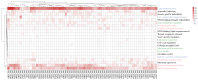
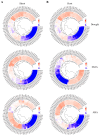
Methods: Using Arabidopsis CCCH proteins as references, we identified the CCCH gene family in rice and analyzed the physicochemical properties, subcellular localization, conserved structures, phylogeny, cis-regulatory elements, synteny analysis, spatiotemporal expression patterns, and expression patterns under drought, ABA, and MeJA treatments for the identified CCCH family members.
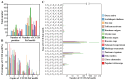
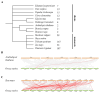
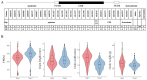
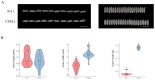
Results: The results showed that the rice CCCH family comprises 73 members, which are unevenly distributed across the 12 chromosomes. Phylogenetic analysis classified them into 11 subfamilies. Subcellular localization indicated that most members are localized in the nucleus. The upstream regions of CCCH promoters contain a large number of cis-regulatory elements related to plant hormones and biotic stress responses. Most genes respond to drought, abscisic acid (ABA), and methyl jasmonate (MeJA) treatments. OsC3H36 was highly expressed under drought, ABA, and MeJA treatments. Haplotype analysis of this gene revealed two major allelic variants (H1 and H2), with H1 predominantly found in japonica rice and associated with increased grain width and 1000-grain weight. Functional validation using a chromosome segment substitution line (CSSL1) confirmed these findings.
Conclusions: CCCH genes play important roles in rice growth, development, and stress responses. Additionally, we validated that OsC3H36 is associated with rice grain width and 1000-grain weight.
Keywords: CCCH gene family; gene expression; haplotype analysis; phenotypic analysis; rice.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

References
-
- Cheng X., Cao J., Gao C., Gao W., Yan S., Yao H., Xu K., Liu X., Xu D., Pan X., et al. Identification of the Wheat C3H Gene Family and Expression Analysis of Candidates Associated with Seed Dormancy and Germination. Plant Physiol. Biochem. 2020;156:524–537. doi: 10.1016/j.plaphy.2020.09.032. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

