Whole-Genome Insights into the Genetic Basis of Conformation Traits in German Black Pied (DSN) Cattle
- PMID: 40282405
- PMCID: PMC12027004
- DOI: 10.3390/genes16040445
Whole-Genome Insights into the Genetic Basis of Conformation Traits in German Black Pied (DSN) Cattle
Abstract
Background: The German Black Pied Dairy (DSN) cattle is an endangered dual-purpose breed considered an ancestor of the modern Holstein population. DSN is known for its high milk yield, favorable milk composition, and good meat quality. Maintaining a functional body structure is essential for ensuring sustained performance across multiple lactations in dual-purpose breeds like DSN. This study aims to identify candidate genes and genetic regions associated with conformation traits in DSN cattle through genome-wide association studies (GWAS).
Methods: The analysis utilized imputed whole-genome sequencing data of 1852 DSN cows with conformation data for 19 linear traits and four composite scores derived from these traits. GWAS was performed using linear mixed models.
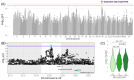
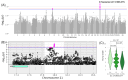
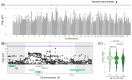
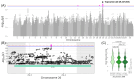
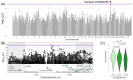
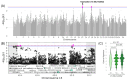
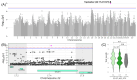
Results: In total, we identified 118 sequence variants distributed across 24 quantitative trait locus (QTL) regions comprising 74 positional candidate genes. Among the most significant findings were variants associated with "Rump width" on chromosome 21 and "Teat length" on chromosome 22, with AGBL1 and SRGAP3 identified as the most likely candidate genes. Additionally, a QTL region on chromosome 15 linked to "Central ligament" contained 39 olfactory receptor genes, and a QTL region on chromosome 23 associated with "Hock quality" included eight immune-related genes, notably, BOLA and TRIM family members.
Conclusions: Selective breeding for favorable alleles of the investigated conformation traits may contribute to DSN's longevity, robustness, and overall resilience. Hence, continuous focus on healthy udders, feet, and legs in herd management contributes to preserving DSN's positive traits while improving conformation.
Keywords: conservation; dual-purpose cattle; endangered cattle breed; genome-wide association study (GWAS); whole-genome sequencing (WGS).
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
Similar articles
-
Genomic insights into growth traits in German Black Pied cattle: a dual-purpose breed at risk.Animal. 2025 Jun;19(6):101540. doi: 10.1016/j.animal.2025.101540. Epub 2025 May 7. Animal. 2025. PMID: 40424954
-
Genetic evaluations for endangered dual-purpose German Black Pied cattle using 50K SNPs, a breed-specific 200K chip, and whole-genome sequencing.J Dairy Sci. 2023 May;106(5):3345-3358. doi: 10.3168/jds.2022-22665. Epub 2023 Apr 5. J Dairy Sci. 2023. PMID: 37028956
-
Design and performance of a bovine 200 k SNP chip developed for endangered German Black Pied cattle (DSN).BMC Genomics. 2021 Dec 18;22(1):905. doi: 10.1186/s12864-021-08237-2. BMC Genomics. 2021. PMID: 34922441 Free PMC article.
-
Genome-wide associations and functional gene analyses for endoparasite resistance in an endangered population of native German Black Pied cattle.BMC Genomics. 2019 Apr 8;20(1):277. doi: 10.1186/s12864-019-5659-4. BMC Genomics. 2019. PMID: 30961534 Free PMC article.
-
Underlying genetic architecture of resistance to mastitis in dairy cattle: A systematic review and gene prioritization analysis of genome-wide association studies.J Dairy Sci. 2023 Jan;106(1):323-351. doi: 10.3168/jds.2022-21923. Epub 2022 Nov 1. J Dairy Sci. 2023. PMID: 36333139
Cited by
-
Genomic Selection for Economically Important Traits in Dual-Purpose Simmental Cattle.Animals (Basel). 2025 Jul 3;15(13):1960. doi: 10.3390/ani15131960. Animals (Basel). 2025. PMID: 40646859 Free PMC article.
References
-
- Bundesanstalt für Landwirtschaft und Ernährung (BLE) Einheimische Nutztierrassen in Deutschland Und Rote Liste Gefährdeter Nutztierrassen 2023. BLE; Bonn, Germany: 2023.
-
- Adler B., Gassan M., Thiele M. Deutsches Schwarzbuntes Niederungsrind—50 Jahre DSN-Genreservezucht in Brandenburg. RBB Rinderproduktion Berlin-Brandenburg GmbH; Groß Kreutz, Germany: 2023.
-
- Rinderzuchtverband Berlin-Brandenburg eG (RZB eG) Zuchtprogramm Für Die Rasse Deutsches Schwarzbuntes Niederungsrind. RZB eG; Groß Kreutz, Germany: 2022.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

