Developmental Validation of DNA Quantitation System, Extended STR Typing Multiplex, and Database Solutions for Panthera leo Genotyping
- PMID: 40283218
- PMCID: PMC12028859
- DOI: 10.3390/life15040664
Developmental Validation of DNA Quantitation System, Extended STR Typing Multiplex, and Database Solutions for Panthera leo Genotyping
Abstract
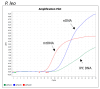
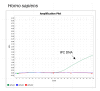
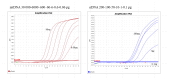
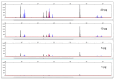
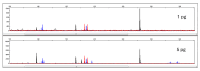
This study describes the development of a species determination/DNA quantification system called Pleo Qplex and an individual identification STR multiplex called Pleo STRplex using Panthera leo samples. Pleo Qplex enables us to measure the quantity of extracted nuclear and mitochondrial DNA and detect the presence of co-purified inhibitors. Pleo STRplex, consisting of seven loci, enables the determination of the DNA profile from a sample of Panthera leo based on the analysis of short tandem repeats (STRs). The Pleo STRplex provides additional loci on top of previously published STR loci in Ptig STRplex and contains a specific STR marker that confirms Panthera leo. An allelic ladder of all STR markers was prepared to enable reliable allele calling. The STR loci can also be used to type the DNA of other members of the genus Panthera. The work on the resulting STR profiles is performed using GenoProof Suite, which offers databasing, matching, and relationship analysis.
Keywords: CITES organism; DNA quantitation; STR multiplex; databasing; forensic genetics; matching analysis; species determination; wildlife.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Molecular Tools for Lynx spp. qPCR Identification and STR-Based Individual Identification of Eurasian Lynx (Lynx lynx) in Forensic Casework.Methods Protoc. 2025 May 2;8(3):47. doi: 10.3390/mps8030047. Methods Protoc. 2025. PMID: 40407474 Free PMC article.
-
Developmental validation of the MiSeq FGx Forensic Genomics System for Targeted Next Generation Sequencing in Forensic DNA Casework and Database Laboratories.Forensic Sci Int Genet. 2017 May;28:52-70. doi: 10.1016/j.fsigen.2017.01.011. Epub 2017 Jan 27. Forensic Sci Int Genet. 2017. PMID: 28171784
-
Development and validation of a novel 133-plex forensic STR panel (52 STRs and 81 Y-STRs) using single-end 400 bp massive parallel sequencing.Int J Legal Med. 2022 Mar;136(2):447-464. doi: 10.1007/s00414-021-02738-1. Epub 2021 Nov 6. Int J Legal Med. 2022. PMID: 34741666
-
Y-Short Tandem Repeat Multiplex Systems - Y-PLEX™ 6 and Y-PLEX™ 5.Forensic Sci Rev. 2003 Jul;15(2):115-35. Forensic Sci Rev. 2003. PMID: 26256728 Review.
-
Biology and Genetics of New Autosomal STR Loci Useful for Forensic DNA Analysis.Forensic Sci Rev. 2012 Jan;24(1):15-26. Forensic Sci Rev. 2012. PMID: 26231356 Review.
References
-
- Brugière D., Chardonnet B., Scholte P. Large-scale extinction of large carnivores (lion Panthera leo, cheetah Acinonyx jubatus and wild dog Lycaon pictus) in protected areas of West and Central Africa. Trop. Conserv. Sci. 2015;8:513–527. doi: 10.1177/194008291500800215. - DOI
-
- Johanisová L., Mauerhofer V. Assessing trophy hunting in South Africa by comparing hunting and exporting databases. J. Nat. Conserv. 2023;72:126363. doi: 10.1016/j.jnc.2023.126363. - DOI
-
- Everatt K., Kokes R., Lopez Pereira C. Evidence of a further emerging threat to lion conservation; targeted poaching for body parts. Biodivers. Conserv. 2019;28:4099–4114. doi: 10.1007/s10531-019-01866-w. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources

