Light-Mediated Population Dynamics of Picocyanobacteria Shaping the Diurnal Patterns of Microbial Communities in an Atoll Lagoon
- PMID: 40284564
- PMCID: PMC12029148
- DOI: 10.3390/microorganisms13040727
Light-Mediated Population Dynamics of Picocyanobacteria Shaping the Diurnal Patterns of Microbial Communities in an Atoll Lagoon
Abstract
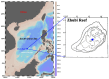
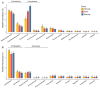
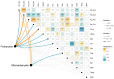
The diurnal cycle of light significantly impacts microbes, making diurnal investigations crucial for understanding microbial communities. Zhubi Reef is known to harbor exceptionally rich biodiversity, with both zooplankton and seawater properties demonstrating diurnal patterns. However, microbial community structures and their potential diurnal dynamics remain largely unexplored. This study is the first to utilize flow cytometry and high-throughput sequencing to investigate prokaryotic and microeukaryotic communities in the Zhubi lagoon, focusing on diurnal variations under different light intensities. The picophytoplankton cell abundance and the microbial community structures both exhibit clear diurnal variations. Light is identified as the primary driver of diurnal variations in the picophytoplankton cell abundance. The diurnal variation in microbial community diversity is driven by changes in the cell abundance of two dominant picocyanobacterial groups. Our findings reveal the diurnal variation in microbial community structures is mediated by the light-driven fluctuation of dominant cyanobacterial populations, and the diurnal variation patterns of specific populations may vary with habitats and sampling timepoints. This research provides valuable insights into the microbial community structure within the Zhubi lagoon.
Keywords: atoll lagoon; diurnal patterns; flow cytometry; high-throughput sequencing; light; microbial community; picocyanobacteria.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




References
-
- Morimoto D., Šulčius S., Tominaga K., Yoshida T. Predetermined clockwork microbial worlds: Current understanding of aquatic microbial diel response from model systems to complex environments. Adv. Appl. Microbiol. 2020;113:163–191. - PubMed
-
- Poretsky R.S., Hewson I., Sun S., Allen A.E., Zehr J.P., Moran M.A. Comparative day/night metatranscriptomic analysis of microbial communities in the North Pacific subtropical gyre. Environ. Microbiol. 2009;11:1358–1375. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

