Multilocus Variable-Number Tandem-Repeat Analysis as an Investigation Tool in Cryptosporidium parvum Outbreaks in Finland and Sweden in 2022
- PMID: 40284657
- PMCID: PMC12029223
- DOI: 10.3390/microorganisms13040821
Multilocus Variable-Number Tandem-Repeat Analysis as an Investigation Tool in Cryptosporidium parvum Outbreaks in Finland and Sweden in 2022
Abstract
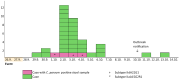
Cryptosporidium is a significant cause of foodborne outbreaks. The 60 kDa glycoprotein gene (gp60) is most often used for subtyping Cryptosporidium species but is not always sufficient for defining clusters and infections sources. The Multilocus Variable-Number Tandem-Repeat Analysis (MLVA) scheme has been developed to better differentiate between subtypes. A cryptosporidiosis outbreak, with 35 cases, was detected in Finland in September 2022. At the same time, in Sweden, three cryptosporidiosis outbreaks, with 107 cases, were detected, leading to international collaboration. In both countries, salad mixes were suspected as being the outbreak source. In the Finnish outbreak, the suspected salad mixes were produced in Sweden. In the Swedish outbreaks, salad mixes from two different producers were suspected. Twenty-nine patient samples which were positive for Cryptosporidium parvum (11 from Finland and 18 from Sweden) were sent for MLVA. The Finnish outbreak samples had different gp60 subtypes and MLVA profiles compared to the Swedish samples. In our investigation, MLVA differentiated C. parvum subtypes in more detail than gp60 typing. MLVA suggested no connection between the Finnish and Swedish outbreaks. A traceback investigation supported this conclusion. To detect outbreaks and identify infection sources, the timely subtyping of patient samples is crucial and should be implemented in routine surveillance and outbreak investigations.
Keywords: MLVA scheme; cohort study; cryptosporidiosis; disease outbreaks; epidemiology; foodborne; molecular typing; subtype.
Conflict of interest statement
W.K. and A.C.G.-P. are fellows of the ECDC Fellowship Programme, supported financially by the European Centre for Disease Prevention and Control. The views and opinions expressed herein do not state or reflect those of the ECDC. The ECDC is not responsible for the data and information collation and analysis and cannot be held liable for the conclusions or opinions drawn. All of the other authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures
References
-
- Checkley W., White A.C., Jr., Jaganath D., Arrowood M.J., Chalmers R.M., Chen X.M., Fayer R., Griffiths J.K., Guerrant R.L., Hedstrom L., et al. A review of the global burden, novel diagnostics, therapeutics, and vaccine targets for cryptosporidium. Lancet Infect. Dis. 2015;15:85–94. doi: 10.1016/S1473-3099(14)70772-8. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

