Staphylococcus aureus Biofilm-Associated Infections: Have We Found a Clinically Relevant Target?
- PMID: 40284688
- PMCID: PMC12029350
- DOI: 10.3390/microorganisms13040852
Staphylococcus aureus Biofilm-Associated Infections: Have We Found a Clinically Relevant Target?
Abstract
Staphylococcus aureus is one of the most diverse bacterial pathogens. This is reflected in its ability to cause a wide array of infections and in genotypic and phenotypic differences between clinical isolates that extend beyond their antibiotic resistance status. Many S. aureus infections, including those involving indwelling medical devices, are therapeutically defined by the formation of a biofilm. This is reflected in the number of reports focusing on S. aureus biofilm formation and biofilm-associated infections. These infections are characterized by a level of intrinsic resistance that compromises conventional antibiotic therapy irrespective of acquired resistance, suggesting that an inhibitor of biofilm formation would have tremendous clinical value. Many reports have described large-scale screens aimed at identifying compounds that limit S. aureus biofilm formation, but relatively few examined whether the limitation was sufficient to overcome this intrinsic resistance. Similarly, while many of these reports examined the impact of putative inhibitors on S. aureus phenotypes, very few took a focused approach to identify and optimize an effective inhibitor of specific biofilm-associated targets. Such approaches are dependent on validating a target, hopefully one that is not restricted by the diversity of S. aureus as a bacterial pathogen. Rigorous biological validation of such a target would allow investigators to virtually screen vast chemical libraries to identify potential inhibitors that warrant further investigation based on their predicted function. Here, we summarize reports describing S. aureus regulatory loci implicated in biofilm formation to assess whether they are viable targets for the development of an anti-biofilm therapeutic strategy with an emphasis on whether sarA has been sufficiently validated to warrant consideration in this important clinical context.
Keywords: Staphylococcus aureus; agr; biofilm; osteomyelitis; protease; regulation; sarA; sigB; xerC.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
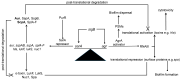
Similar articles
-
The ability of sarA to limit protease production plays a key role in the pathogenesis of Staphylococcus aureus osteomyelitis irrespective of the functional status of agr.Infect Immun. 2025 Jan 31;93(1):e0047324. doi: 10.1128/iai.00473-24. Epub 2024 Nov 29. Infect Immun. 2025. PMID: 39611695 Free PMC article.
-
The Black Book of Psychotropic Dosing and Monitoring.Psychopharmacol Bull. 2024 Jul 8;54(3):8-59. Psychopharmacol Bull. 2024. PMID: 38993656 Free PMC article. Review.
-
Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19.Cochrane Database Syst Rev. 2022 May 20;5(5):CD013665. doi: 10.1002/14651858.CD013665.pub3. Cochrane Database Syst Rev. 2022. PMID: 35593186 Free PMC article.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2017 Dec 22;12(12):CD011535. doi: 10.1002/14651858.CD011535.pub2. Cochrane Database Syst Rev. 2017. Update in: Cochrane Database Syst Rev. 2020 Jan 9;1:CD011535. doi: 10.1002/14651858.CD011535.pub3. PMID: 29271481 Free PMC article. Updated.
-
A rapid and systematic review of the clinical effectiveness and cost-effectiveness of paclitaxel, docetaxel, gemcitabine and vinorelbine in non-small-cell lung cancer.Health Technol Assess. 2001;5(32):1-195. doi: 10.3310/hta5320. Health Technol Assess. 2001. PMID: 12065068
Cited by
-
Staphylococci in Livestock: Molecular Epidemiology, Antimicrobial Resistance, and Translational Strategies for One Health Protection.Vet Sci. 2025 Aug 13;12(8):757. doi: 10.3390/vetsci12080757. Vet Sci. 2025. PMID: 40872707 Free PMC article. Review.
References
-
- Coifu O., Moser C., Jensen P.Ø., HØiby N. Tolerance and resistance of microbial biofilms. Nat. Rev. Microbiol. 2022;20:621–635. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

